Metagenomic DNA sequencing to quantify Mycobacterium tuberculosis DNA and diagnose tuberculosis
- PMID: 36216964
- PMCID: PMC9551046
- DOI: 10.1038/s41598-022-21244-x
Metagenomic DNA sequencing to quantify Mycobacterium tuberculosis DNA and diagnose tuberculosis
Abstract
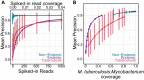
Tuberculosis (TB) remains a significant cause of mortality worldwide. Metagenomic next-generation sequencing has the potential to reveal biomarkers of active disease, identify coinfection, and improve detection for sputum-scarce or culture-negative cases. We conducted a large-scale comparative study of 428 plasma, urine, and oral swab samples from 334 individuals from TB endemic and non-endemic regions to evaluate the utility of a shotgun metagenomic DNA sequencing assay for tuberculosis diagnosis. We found that the composition of the control population had a strong impact on the measured performance of the diagnostic test: the use of a control population composed of individuals from a TB non-endemic region led to a test with nearly 100% specificity and sensitivity, whereas a control group composed of individuals from TB endemic regions exhibited a high background of nontuberculous mycobacterial DNA, limiting the diagnostic performance of the test. Using mathematical modeling and quantitative comparisons to matched qPCR data, we found that the burden of Mycobacterium tuberculosis DNA constitutes a very small fraction (0.04 or less) of the total abundance of DNA originating from mycobacteria in samples from TB endemic regions. Our findings suggest that the utility of a minimally invasive metagenomic sequencing assay for pulmonary tuberculosis diagnostics is limited by the low burden of M. tuberculosis and an overwhelming biological background of nontuberculous mycobacterial DNA.
© 2022. The Author(s).
Conflict of interest statement
I.D.V. has received research grants from the Bill and Melinda Gates Foundation, the National Institutes of Health, and the Rainin Foundation. P.B. has received research grants from the National Science Foundation. I.D.V. and P.B. are inventors on the patent US-2020-0048713-A1 titled “Methods of Detecting Cell-Free DNA in Biological Samples”. The rights to the patent were licensed by Eurofins through Cornell University. I.D.V. is a Member of the Advisory Board and has stock in Karius Inc. A.C., O.M., L.D.K., J.L., P.K., A.A., J.C., C.M.B., A.C., and A.S. declare no potential conflict of interest.
Figures



References
-
- Bowness R, et al. The relationship between Mycobacterium tuberculosis MGIT time to positivity and cfu in sputum samples demonstrates changing bacterial phenotypes potentially reflecting the impact of chemotherapy on critical sub-populations. J. Antimicrob. Chemother. 2015;70:448–455. doi: 10.1093/jac/dku415. - DOI - PubMed

