Impaired TFEB activation and mitophagy as a cause of PPP3/calcineurin inhibitor-induced pancreatic β-cell dysfunction
- PMID: 36217215
- PMCID: PMC10240995
- DOI: 10.1080/15548627.2022.2132686
Impaired TFEB activation and mitophagy as a cause of PPP3/calcineurin inhibitor-induced pancreatic β-cell dysfunction
Abstract
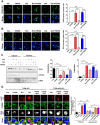
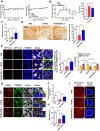
Macroautophagy/autophagy or mitophagy plays crucial roles in the maintenance of pancreatic β-cell function. PPP3/calcineurin can modulate the activity of TFEB, a master regulator of lysosomal biogenesis and autophagy gene expression, through dephosphorylation. We studied whether PPP3/calcineurin inhibitors can affect the mitophagy of pancreatic β-cells and pancreatic β-cell function employing FK506, an immunosuppressive drug against graft rejection. FK506 suppressed rotenone- or oligomycin+antimycin-A-induced mitophagy measured by Mito-Keima localization in acidic lysosomes or RFP-LC3 puncta colocalized with TOMM20 in INS-1 insulinoma cells. FK506 diminished nuclear translocation of TFEB after treatment with rotenone or oligomycin+antimycin A. Forced TFEB nuclear translocation by a constitutively active TFEB mutant transfection restored impaired mitophagy by FK506, suggesting the role of decreased TFEB nuclear translocation in FK506-mediated mitophagy impairment. Probably due to reduced mitophagy, recovery of mitochondrial potential or quenching of mitochondrial ROS after removal of rotenone or oligomycin+antimycin A was delayed by FK506. Mitochondrial oxygen consumption was reduced by FK506, indicating reduced mitochondrial function by FK506. Likely due to mitochondrial dysfunction, insulin release from INS-1 cells was reduced by FK506 in vitro. FK506 treatment also reduced insulin release and impaired glucose tolerance in vivo, which was associated with decreased mitophagy and mitochondrial COX activity in pancreatic islets. FK506-induced mitochondrial dysfunction and glucose intolerance were ameliorated by an autophagy enhancer activating TFEB. These results suggest that diminished mitophagy and consequent mitochondrial dysfunction of pancreatic β-cells contribute to FK506-induced β-cell dysfunction or glucose intolerance, and autophagy enhancement could be a therapeutic modality against post-transplantation diabetes mellitus caused by PPP3/calcineurin inhibitors.
Keywords: Calcineurin; TFEB; mitophagy; pancreatic β-cell; post-transplantation diabetes mellitus.
Conflict of interest statement
M.-S. L. is the CEO of LysoTech, Inc.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
