EZ Clear for simple, rapid, and robust mouse whole organ clearing
- PMID: 36218247
- PMCID: PMC9555867
- DOI: 10.7554/eLife.77419
EZ Clear for simple, rapid, and robust mouse whole organ clearing
Abstract
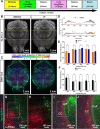
Tissue clearing for whole organ cell profiling has revolutionized biology and imaging for exploration of organs in three-dimensional space without compromising tissue architecture. But complicated, laborious procedures, or expensive equipment, as well as the use of hazardous, organic solvents prevent the widespread adoption of these methods. Here, we report a simple and rapid tissue clearing method, EZ Clear, that can clear whole adult mouse organs in 48 hr in just three simple steps. Samples stay at room temperature and remain hydrated throughout the clearing process, preserving endogenous and synthetic fluorescence, without altering sample size. After wholemount clearing and imaging, samples processed with EZ Clear can be subjected to downstream applications, such as tissue embedding and cryosectioning followed by standard histology or immunofluorescent staining without loss of fluorescence signal from endogenous or synthetic reporters. Furthermore, we demonstrate that wholemount adult mouse brains processed with EZ Clear can be successfully immunolabeled for fluorescent imaging while still retaining signal from endogenous fluorescent reporters. Overall, the simplicity, speed, and flexibility of EZ Clear make it easy to adapt and implement in diverse imaging modalities in biomedical research.
Keywords: 3D imaging; cancer biology; developmental biology; mouse; neuroscience; tissue clearing.
© 2022, Hsu et al.
Conflict of interest statement
CH, JC, JK, WT, TR, CF, MD, JW No competing interests declared
Figures








References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

