Noncanonical Mismatch Repair Protein NucS Modulates the Emergence of Antibiotic Resistance in Mycobacterium abscessus
- PMID: 36219122
- PMCID: PMC9769700
- DOI: 10.1128/spectrum.02228-22
Noncanonical Mismatch Repair Protein NucS Modulates the Emergence of Antibiotic Resistance in Mycobacterium abscessus
Abstract
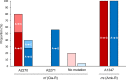
NucS/EndoMS-dependent noncanonical mismatch repair (MMR) ensures the stability of genomic DNA in mycobacteria and acts as a guardian of the genome by preventing the accumulation of point mutations. In order to address whether the inactivation of noncanonical MMR could increase the acquisition of drug resistance by mutation, a ΔnucS strain was constructed and explored in the emerging pathogen Mycobacterium abscessus. Deletion of nucS resulted in a mutator phenotype with increased acquisition of resistance to macrolides and aminoglycosides, the two main groups of antimycobacterial agents for M. abscessus treatment, and also to second-line drugs such as fluoroquinolones. Inactivation of the noncanonical MMR in M. abscessus led to increases of 10- to 22-fold in the appearance of spontaneous mutants resistant to the macrolide clarithromycin and the aminoglycosides amikacin, gentamicin, and apramycin, compared with the wild-type strain. Furthermore, emergence of fluoroquinolone (ciprofloxacin) resistance was detected in a nucS-deficient strain but not in a wild-type M. abscessus strain. Acquired drug resistance to macrolides and aminoglycosides was analyzed through sequencing of the 23S rRNA gene rrl and the 16S rRNA gene rrs from independent drug-resistant colonies of both strains. When the acquisition of clarithromycin resistance was examined, a different mutational profile was detected in the M. abscessus ΔnucS strain compared with the wild-type one. To summarize, M. abscessus requires the NucS-dependent noncanonical MMR pathway to prevent the emergence of drug-resistant isolates by mutation. To our knowledge, this is the first report that reveals the role of NucS in a human pathogen, and these findings have potential implications for the treatment of M. abscessus infections. IMPORTANCE Chronic infections caused by M. abscessus are an emerging challenge in public health, posing a substantial health and economic burden, especially in patients with cystic fibrosis. Treatment of M. abscessus infections with antibiotics is particularly challenging, as its complex drug resistance mechanisms, including constitutive resistance through DNA mutation, lead to high rates of treatment failure. To decipher the evolution of antibiotic resistance in M. abscessus, we studied NucS-dependent noncanonical MMR, a unique DNA repair pathway involved in genomic maintenance. Inactivation of NucS is linked to the increase of DNA mutations (hypermutation), which can confer drug resistance. Our analysis detected increased acquisition of mutations conferring resistance to first-line and second-line antibiotics. We believe that this study will improve the knowledge of how this pathogen could evolve into an untreatable infectious agent, and it uncovers a role for hypermutators in chronic infectious diseases under antibiotic pressure.
Keywords: DNA repair; EndoMS; Mycobacterium abscessus; NucS; acquired resistance; aminoglycosides; antibiotic resistance; drug resistance; macrolides; mismatch repair; mutation; noncanonical mismatch repair.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Bryant JM, Grogono DM, Rodriguez-Rincon D, Everall I, Brown KP, Moreno P, Verma D, Hill E, Drijkoningen J, Gilligan P, Esther CR, Noone PG, Giddings O, Bell SC, Thomson R, Wainwright CE, Coulter C, Pandey S, Wood ME, Stockwell RE, Ramsay KA, Sherrard LJ, Kidd TJ, Jabbour N, Johnson GR, Knibbs LD, Morawska L, Sly PD, Jones A, Bilton D, Laurenson I, Ruddy M, Bourke S, Bowler ICJW, Chapman SJ, Clayton A, Cullen M, Dempsey O, Denton M, Desai M, Drew RJ, Edenborough F, Evans J, Folb J, Daniels T, Humphrey H, Isalska B, Jensen-Fangel S, Jönsson B, Jones AM, et al. . 2016. Emergence and spread of a human-transmissible multidrug-resistant nontuberculous mycobacterium. Science 354:751–757. doi:10.1126/science.aaf8156. - DOI - PMC - PubMed
-
- Bronson RA, Gupta C, Manson AL, Nguyen JA, Bahadirli-Talbott A, Parrish NM, Earl AM, Cohen KA. 2021. Global phylogenomic analyses of Mycobacterium abscessus provide context for non cystic fibrosis infections and the evolution of antibiotic resistance. Nat Commun 12. doi:10.1038/s41467-021-25484-9. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

