Tensor denoising of multidimensional MRI data
- PMID: 36219475
- PMCID: PMC10092037
- DOI: 10.1002/mrm.29478
Tensor denoising of multidimensional MRI data
Abstract
Purpose: To develop a denoising strategy leveraging redundancy in high-dimensional data.
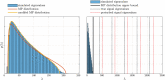
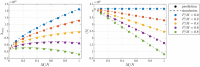
Theory and methods: The SNR fundamentally limits the information accessible by MRI. This limitation has been addressed by a host of denoising techniques, recently including the so-called MPPCA: principal component analysis of the signal followed by automated rank estimation, exploiting the Marchenko-Pastur distribution of noise singular values. Operating on matrices comprised of data patches, this popular approach objectively identifies noise components and, ideally, allows noise to be removed without introducing artifacts such as image blurring, or nonlocal averaging. The MPPCA rank estimation, however, relies on a large number of noise singular values relative to the number of signal components to avoid such ill effects. This condition is unlikely to be met when data patches and therefore matrices are small, for example due to spatially varying noise. Here, we introduce tensor MPPCA (tMPPCA) for the purpose of denoising multidimensional data, such as from multicontrast acquisitions. Rather than combining dimensions in matrices, tMPPCA uses each dimension of the multidimensional data's inherent tensor-structure to better characterize noise, and to recursively estimate signal components.
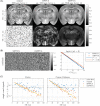
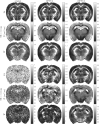
Results: Relative to matrix-based MPPCA, tMPPCA requires no additional assumptions, and comparing the two in a numerical phantom and a multi-TE diffusion MRI data set, tMPPCA dramatically improves denoising performance. This is particularly true for small data patches, suggesting that tMPPCA can be especially beneficial in such cases.
Conclusions: The MPPCA denoising technique can be extended to high-dimensional data with improved performance for smaller patch sizes.
Keywords: denoising; diffusion; principal component analysis; random matrix theory.
© 2022 The Authors. Magnetic Resonance in Medicine published by Wiley Periodicals LLC on behalf of International Society for Magnetic Resonance in Medicine.
Figures
References
-
- Jones DK. Diffusion MRI Theory, Methods, and Applications. Oxford University Press; 2010.
-
- Jones DK. Precision and accuracy in diffusion tensor magnetic resonance imaging. Top Magn Reson Imaging. 2010;21:87‐99. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

