Mutually exclusive genetic interactions and gene essentiality shape the genomic landscape of primary melanoma
- PMID: 36219477
- PMCID: PMC10098817
- DOI: 10.1002/path.6019
Mutually exclusive genetic interactions and gene essentiality shape the genomic landscape of primary melanoma
Abstract
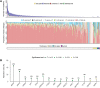
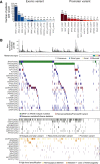
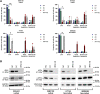
Melanoma is a heterogenous malignancy with an unpredictable clinical course. Most patients who present in the clinic are diagnosed with primary melanoma, yet large-scale sequencing efforts have focused primarily on metastatic disease. In this study we sequence-profiled 524 American Joint Committee on Cancer Stage I-III primary tumours. Our analysis of these data reveals recurrent driver mutations, mutually exclusive genetic interactions, where two genes were never or rarely co-mutated, and an absence of co-occurring genetic events. Further, we intersected copy number calls from our primary melanoma data with whole-genome CRISPR screening data to identify the transcription factor interferon regulatory factor 4 (IRF4) as a melanoma-associated dependency. © 2022 The Authors. The Journal of Pathology published by John Wiley & Sons Ltd on behalf of The Pathological Society of Great Britain and Ireland.
Keywords: CRISPR screening; driver genes; gene essentiality; genetic epistasis; genome sequencing; genomic landscape; interferon regulatory factor 4 (IRF4); melanoma; primary cancer; somatic mutation.
© 2022 The Authors. The Journal of Pathology published by John Wiley & Sons Ltd on behalf of The Pathological Society of Great Britain and Ireland.
Figures


References
-
- Elder DE, Bastian BC, Cree IA, et al. The 2018 World Health Organization classification of cutaneous, mucosal, and uveal melanoma: detailed analysis of 9 distinct subtypes defined by their evolutionary pathway. Arch Pathol Lab Med 2020; 144: 500–522. - PubMed
-
- Clark WH Jr, From L, Bernardino EA, et al. The histogenesis and biologic behavior of primary human malignant melanomas of the skin. Cancer Res 1969; 29: 705–727. - PubMed
-
- Armstrong BK, Kricker A. The epidemiology of UV induced skin cancer. J Photochem Photobiol B 2001; 63: 8–18. - PubMed
-
- Healy E, Flannagan N, Ray A, et al. Melanocortin‐1‐receptor gene and sun sensitivity in individuals without red hair. Lancet 2000; 355: 1072–1073. - PubMed
-
- Valverde P, Healy E, Jackson I, et al. Variants of the melanocyte‐stimulating hormone receptor gene are associated with red hair and fair skin in humans. Nat Genet 1995; 11: 328–330. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

