Genetic Mapping of Multiple Traits Identifies Novel Genes for Adiposity, Lipids, and Insulin Secretory Capacity in Outbred Rats
- PMID: 36219827
- PMCID: PMC9797320
- DOI: 10.2337/db22-0252
Genetic Mapping of Multiple Traits Identifies Novel Genes for Adiposity, Lipids, and Insulin Secretory Capacity in Outbred Rats
Abstract
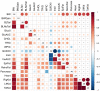
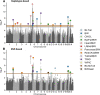
Despite the successes of human genome-wide association studies, the causal genes underlying most metabolic traits remain unclear. We used outbred heterogeneous stock (HS) rats, coupled with expression data and mediation analysis, to identify quantitative trait loci (QTLs) and candidate gene mediators for adiposity, glucose tolerance, serum lipids, and other metabolic traits. Physiological traits were measured in 1,519 male HS rats, with liver and adipose transcriptomes measured in >410 rats. Genotypes were imputed from low-coverage whole-genome sequencing. Linear mixed models were used to detect physiological and expression QTLs (pQTLs and eQTLs, respectively), using both single nucleotide polymorphism (SNP)- and haplotype-based models for pQTL mapping. Genes with cis-eQTLs that overlapped pQTLs were assessed as causal candidates through mediation analysis. We identified 14 SNP-based pQTLs and 19 haplotype-based pQTLs, of which 10 were in common. Using mediation, we identified the following genes as candidate mediators of pQTLs: Grk5 for fat pad weight and serum triglyceride pQTLs on Chr1, Krtcap3 for fat pad weight and serum triglyceride pQTLs on Chr6, Ilrun for a fat pad weight pQTL on Chr20, and Rfx6 for a whole pancreatic insulin content pQTL on Chr20. Furthermore, we verified Grk5 and Ktrcap3 using gene knockdown/out models, thereby shedding light on novel regulators of obesity.
© 2022 by the American Diabetes Association.
Figures




References
-
- Maes HH, Neale MC, Eaves LJ. Genetic and environmental factors in relative body weight and human adiposity. Behav Genet 1997;27:325–351 - PubMed
-
- Klarin D, Damrauer SM, Cho K, et al. .; Global Lipids Genetics Consortium; Myocardial Infarction Genetics (MIGen) Consortium; Geisinger-Regeneron DiscovEHR Collaboration; VA Million Veteran Program . Genetics of blood lipids among ∼300,000 multi-ethnic participants of the Million Veteran Program. Nat Genet 2018;50:1514–1523 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

