Coronavirus Antibody Responses before COVID-19 Pandemic, Africa and Thailand
- PMID: 36220131
- PMCID: PMC9622245
- DOI: 10.3201/eid2811.221041
Coronavirus Antibody Responses before COVID-19 Pandemic, Africa and Thailand
Abstract
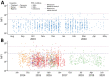
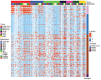
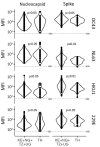
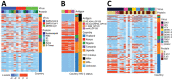
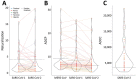
Prior immune responses to coronaviruses might affect human SARS-CoV-2 response. We screened 2,565 serum and plasma samples collected from 2013 through early 2020, before the COVID-19 pandemic began, from 2,250 persons in 4 countries in Africa (Kenya, Nigeria, Tanzania, and Uganda) and in Thailand, including persons living with HIV-1. We detected IgG responses to SARS-CoV-2 spike (S) subunit 2 protein in 1.8% of participants. Profiling against 23 coronavirus antigens revealed that responses to S, subunit 2, or subunit 1 proteins were significantly more frequent than responses to the receptor-binding domain, S-Trimer, or nucleocapsid proteins (p<0.0001). We observed similar responses in persons with or without HIV-1. Among all coronavirus antigens tested, SARS-CoV-2, SARS-CoV-1, and Middle East respiratory syndrome coronavirus antibody responses were much higher in participants from Africa than in participants from Thailand (p<0.01). We noted less pronounced differences for endemic coronaviruses. Serosurveys could affect vaccine and monoclonal antibody distribution across global populations.
Keywords: Africa; COVID-19; HIV-1; SARS; SARS-CoV-2; Thailand; coronavirus; coronavirus disease; respiratory infections; serosurvey; severe acute respiratory syndrome coronavirus 2; viruses; zoonoses.
Figures


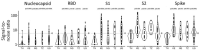
References
-
- Gill CJ, Mwananyanda L, MacLeod W, Kwenda G, Pieciak R, Etter L, et al. Sustained high prevalence of COVID-19 deaths from a systematic post-mortem study in Lusaka, Zambia: one year later. medRxiv. 2022. 10.1101/2022.03.08.22272087 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

