A simple SEIR-V model to estimate COVID-19 prevalence and predict SARS-CoV-2 transmission using wastewater-based surveillance data
- PMID: 36220466
- PMCID: PMC9547654
- DOI: 10.1016/j.scitotenv.2022.159326
A simple SEIR-V model to estimate COVID-19 prevalence and predict SARS-CoV-2 transmission using wastewater-based surveillance data
Abstract
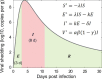
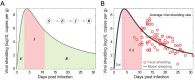
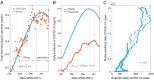
Wastewater-based surveillance (WBS) has been widely used as a public health tool to monitor SARS-CoV-2 transmission. However, epidemiological inference from WBS data remains understudied and limits its application. In this study, we have established a quantitative framework to estimate COVID-19 prevalence and predict SARS-CoV-2 transmission through integrating WBS data into an SEIR-V model. We conceptually divide the individual-level viral shedding course into exposed, infectious, and recovery phases as an analogy to the compartments in a population-level SEIR model. We demonstrated that the effect of temperature on viral losses in the sewer can be straightforwardly incorporated in our framework. Using WBS data from the second wave of the pandemic (Oct 02, 2020-Jan 25, 2021) in the Greater Boston area, we showed that the SEIR-V model successfully recapitulates the temporal dynamics of viral load in wastewater and predicts the true number of cases peaked earlier and higher than the number of reported cases by 6-16 days and 8.3-10.2 folds (R = 0.93). This work showcases a simple yet effective method to bridge WBS and quantitative epidemiological modeling to estimate the prevalence and transmission of SARS-CoV-2 in the sewershed, which could facilitate the application of wastewater surveillance of infectious diseases for epidemiological inference and inform public health actions.
Keywords: Epidemic model; SARS-CoV-2; SEIR-V model; Temperature; Wastewater-based epidemiology.
Copyright © 2022 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

Update of
-
A simple SEIR-V model to estimate COVID-19 prevalence and predict SARS-CoV-2 transmission using wastewater-based surveillance data.medRxiv [Preprint]. 2022 Jul 18:2022.07.17.22277721. doi: 10.1101/2022.07.17.22277721. medRxiv. 2022. Update in: Sci Total Environ. 2023 Jan 20;857(Pt 1):159326. doi: 10.1016/j.scitotenv.2022.159326. PMID: 35898336 Free PMC article. Updated. Preprint.
References
-
- Bˇehrádek J. Temperature coefficients in biology. Biol. Rev. 1930;5(1):30–58.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

