Arylcoumarin perturbs SARS-CoV-2 pathogenesis by targeting the S-protein/ACE2 interaction
- PMID: 36220880
- PMCID: PMC9552724
- DOI: 10.1038/s41598-022-20759-7
Arylcoumarin perturbs SARS-CoV-2 pathogenesis by targeting the S-protein/ACE2 interaction
Abstract
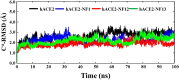
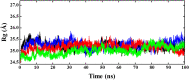
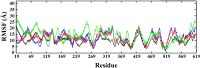
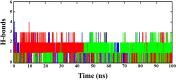
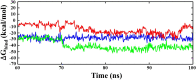
The vaccination drive against COVID-19 worldwide was quite successful. However, the second wave of infections was even more disastrous. There was a rapid increase in reinfections and human deaths due to the appearance of new SARS-CoV-2 variants. The viral genome mutations in the variants were acquired while passing through different human hosts that could escape antibodies in convalescent or vaccinated individuals. The treatment was based on oxygen supplements and supportive protocols due to the lack of a specific drug. In this study, we identified three lead inhibitors of arylated coumarin derivatives 4,6,8-tri(naphthalen-2-yl)-2H-chromen-2-one (NF1), 8-(4-hydroxyphenyl)-4,6-di(naphthalen-2-yl)-2H-chromen-2-one (NF12) and 8-(4-hydroxyphenyl)-3,6-di(naphthalen-2-yl)-2H-chromen-2-one (NF-13) that showed higher binding affinity towards the junction of SARS-CoV-2 spike glycoprotein (S-protein) and human angiotensin-converting enzyme 2 (ACE2) receptor. Using molecular docking analysis, we identified the putative binding sites of these potent inhibitors. Notably, molecular dynamics (MD) simulation and MM-PBSA studies confirmed that these inhibitors have the potential ability to bind Spike-protein/ACE2 protein complex with minimal energy. Further, the two major concerns are an adaptive mutation of spike proteins- N501Y and D614G which displayed strong affinity towards NF-13 in docking analysis. Additionally, in vitro and in vivo studies are required to confirm the above findings and develop the inhibitors as potential drugs against SARS-CoV-2.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Investigation of nonsynonymous mutations in the spike protein of SARS-CoV-2 and its interaction with the ACE2 receptor by molecular docking and MM/GBSA approach.Comput Biol Med. 2021 Aug;135:104654. doi: 10.1016/j.compbiomed.2021.104654. Epub 2021 Jul 16. Comput Biol Med. 2021. PMID: 34346317 Free PMC article.
-
V367F Mutation in SARS-CoV-2 Spike RBD Emerging during the Early Transmission Phase Enhances Viral Infectivity through Increased Human ACE2 Receptor Binding Affinity.J Virol. 2021 Jul 26;95(16):e0061721. doi: 10.1128/JVI.00617-21. Epub 2021 Jul 26. J Virol. 2021. PMID: 34105996 Free PMC article.
-
Multidisciplinary Approaches Identify Compounds that Bind to Human ACE2 or SARS-CoV-2 Spike Protein as Candidates to Block SARS-CoV-2-ACE2 Receptor Interactions.mBio. 2021 Mar 30;12(2):e03681-20. doi: 10.1128/mBio.03681-20. mBio. 2021. PMID: 33785634 Free PMC article.
-
The SARS-CoV-2 Spike Glycoprotein as a Drug and Vaccine Target: Structural Insights into Its Complexes with ACE2 and Antibodies.Cells. 2020 Oct 22;9(11):2343. doi: 10.3390/cells9112343. Cells. 2020. PMID: 33105869 Free PMC article. Review.
-
SARS-CoV-2 pandemic and research gaps: Understanding SARS-CoV-2 interaction with the ACE2 receptor and implications for therapy.Theranostics. 2020 Jun 12;10(16):7448-7464. doi: 10.7150/thno.48076. eCollection 2020. Theranostics. 2020. PMID: 32642005 Free PMC article. Review.
Cited by
-
Elucidating the binding specificity of interactive compounds targeting ATP-binding cassette subfamily G member 2 (ABCG2).Mol Divers. 2025 Jan 9. doi: 10.1007/s11030-024-11078-2. Online ahead of print. Mol Divers. 2025. PMID: 39786520
-
Antiviral Effect of Piperine on Chikungunya Virus: In Vitro Evidence and In Silico Analysis of E1-E2 Binding.ACS Omega. 2025 Aug 6;10(32):35865-35877. doi: 10.1021/acsomega.5c02814. eCollection 2025 Aug 19. ACS Omega. 2025. PMID: 40852290 Free PMC article.
-
Hydroxyethylamine based analog targets microtubule assembly: an in silico study for anti-cancerous drug development.Sci Rep. 2024 Dec 28;14(1):31381. doi: 10.1038/s41598-024-82823-8. Sci Rep. 2024. PMID: 39732970 Free PMC article.
References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

