Ligand-induced shifts in conformational ensembles that describe transcriptional activation
- PMID: 36222302
- PMCID: PMC9555869
- DOI: 10.7554/eLife.80140
Ligand-induced shifts in conformational ensembles that describe transcriptional activation
Abstract
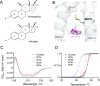
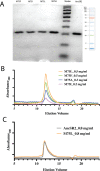
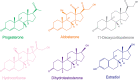
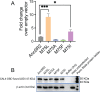
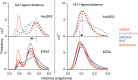
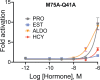
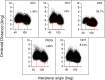
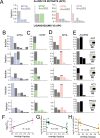
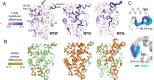
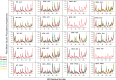
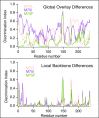
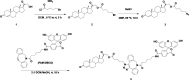
Nuclear receptors function as ligand-regulated transcription factors whose ability to regulate diverse physiological processes is closely linked with conformational changes induced upon ligand binding. Understanding how conformational populations of nuclear receptors are shifted by various ligands could illuminate strategies for the design of synthetic modulators to regulate specific transcriptional programs. Here, we investigate ligand-induced conformational changes using a reconstructed, ancestral nuclear receptor. By making substitutions at a key position, we engineer receptor variants with altered ligand specificities. We combine cellular and biophysical experiments to characterize transcriptional activity, as well as elucidate mechanisms underlying altered transcription in receptor variants. We then use atomistic molecular dynamics (MD) simulations with enhanced sampling to generate ensembles of wildtype and engineered receptors in combination with multiple ligands, followed by conformational analysis and correlation of MD-based predictions with functional ligand profiles. We determine that conformational ensembles accurately describe ligand responses based on observed population shifts. These studies provide a platform which will allow structural characterization of physiologically-relevant conformational ensembles, as well as provide the ability to design and predict transcriptional responses in novel ligands.
Keywords: biochemistry; chemical biology; conformational ensembles; molecular biophysics; molecular dynamics; none; nuclear receptors; protein dynamics; structural biology; transcriptional activation.
© 2022, Khan et al.
Conflict of interest statement
SK, SB, SK, EE, GA, CO No competing interests declared
Figures









Similar articles
-
Improved docking, screening and selectivity prediction for small molecule nuclear receptor modulators using conformational ensembles.J Comput Aided Mol Des. 2010 May;24(5):459-71. doi: 10.1007/s10822-010-9362-4. Epub 2010 May 9. J Comput Aided Mol Des. 2010. PMID: 20455005 Free PMC article.
-
Exploring conformational landscapes and binding mechanisms of convergent evolution for the SARS-CoV-2 spike Omicron variant complexes with the ACE2 receptor using AlphaFold2-based structural ensembles and molecular dynamics simulations.Phys Chem Chem Phys. 2024 Jun 26;26(25):17720-17744. doi: 10.1039/d4cp01372g. Phys Chem Chem Phys. 2024. PMID: 38869513
-
Diverse Native Ensembles Dictate the Differential Functional Responses of Nuclear Receptor Ligand-Binding Domains.J Phys Chem B. 2021 Apr 15;125(14):3546-3555. doi: 10.1021/acs.jpcb.1c00972. Epub 2021 Apr 4. J Phys Chem B. 2021. PMID: 33818099
-
Molecular Dynamics Simulations Combined with Nuclear Magnetic Resonance and/or Small-Angle X-ray Scattering Data for Characterizing Intrinsically Disordered Protein Conformational Ensembles.J Chem Inf Model. 2019 May 28;59(5):1743-1758. doi: 10.1021/acs.jcim.8b00928. Epub 2019 Mar 18. J Chem Inf Model. 2019. PMID: 30840442 Review.
-
Small molecule modulation of nuclear receptor conformational dynamics: implications for function and drug discovery.Mol Pharmacol. 2013 Jan;83(1):1-8. doi: 10.1124/mol.112.079285. Epub 2012 Aug 6. Mol Pharmacol. 2013. PMID: 22869589 Free PMC article. Review.
Cited by
-
Drug resistance and tumor heterogeneity: cells and ensembles.Biophys Rev. 2025 May 31;17(3):759-779. doi: 10.1007/s12551-025-01320-y. eCollection 2025 Jun. Biophys Rev. 2025. PMID: 40727661 Free PMC article. Review.
-
Simulations Reveal Unique Roles for the FXR Hinge in the FXR-RXR Nuclear Receptor Heterodimer.ACS Bio Med Chem Au. 2024 Nov 27;5(1):194-203. doi: 10.1021/acsbiomedchemau.4c00105. eCollection 2025 Feb 19. ACS Bio Med Chem Au. 2024. PMID: 39990948 Free PMC article.
-
Cell phenotypes can be predicted from propensities of protein conformations.Curr Opin Struct Biol. 2023 Dec;83:102722. doi: 10.1016/j.sbi.2023.102722. Epub 2023 Oct 21. Curr Opin Struct Biol. 2023. PMID: 37871498 Free PMC article. Review.
-
Enhanced dynamic coupling in a nuclear receptor underlies ligand activity.J Biol Chem. 2025 Feb;301(2):108081. doi: 10.1016/j.jbc.2024.108081. Epub 2024 Dec 14. J Biol Chem. 2025. PMID: 39675705 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

