Uncovering the promiscuous activity of IL-6 proteins: A multi-dimensional analysis of phylogeny, classification and residue conservation
- PMID: 36222303
- PMCID: PMC9601804
- DOI: 10.1002/pro.4469
Uncovering the promiscuous activity of IL-6 proteins: A multi-dimensional analysis of phylogeny, classification and residue conservation
Abstract
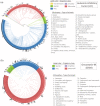
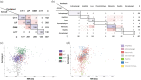
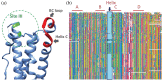
The IL-6 family of cytokines, known for their pleiotropic behavior, share binding to the gp130 receptor for signal transduction with the necessity to bind other receptors. Leukemia inhibitory factor receptor is triggered by the IL-6 family proteins: leukemia inhibitory factor (LIF), oncostatin-M (OSM), cardiotrophin-1 (CT-1), ciliary neurotrophic factor (CNTF), and cardiotrophin-like cytokine factor 1 (CLCF1). Besides the conserved binding sites to the receptor, not much is known in terms of the diversity and characteristics of these proteins in different organisms. Herein, we describe the sequence analysis of LIF, OSM, and CT-1 from several organisms, and m17, a LIF ortholog found in fishes, regarding its phylogenetics, intrinsic properties, and the impact of conserved residues on structural features. Sequences were identified in seven classes of vertebrates, showing high conservation values in binding site III, but protein-dependent results on binding site II. GRAVY, isoelectric point, and molecular weight parameters were relevant to differentiate classes in each protein and to enable, for the first time and with high fidelity, the prediction of both organism class and protein type just using machine learning approaches. OSM sequences from primates showed an increased BC loop when compared to the remaining mammals, which could influence binding to OSM receptor and tune signaling pathways. Overall, this study highlights the potential of sequence diversity analysis to understand IL-6 cytokine family evolution, showing the conservation of function-related motifs and evolution of class and protein-dependent characteristics. Our results could impact future medical treatment of disorders associated with imbalances in these cytokines.
Keywords: IL-6 cytokine family; leukemia inhibitory factor; machine learning; protein evolution; sequence diversity analysis.
© 2022 The Authors. Protein Science published by Wiley Periodicals LLC on behalf of The Protein Society.
Figures





Similar articles
-
Interleukin-6 family of cytokines induced activation of different functional sites expressed by gp130 transducing protein.J Biol Chem. 1996 Jun 21;271(25):14764-72. doi: 10.1074/jbc.271.25.14764. J Biol Chem. 1996. PMID: 8662918
-
Leukemia inhibitory factor (LIF), cardiotrophin-1, and oncostatin M share structural binding determinants in the immunoglobulin-like domain of LIF receptor.J Biol Chem. 2003 Jul 18;278(29):27169-79. doi: 10.1074/jbc.M303168200. Epub 2003 Apr 21. J Biol Chem. 2003. PMID: 12707269
-
Cloning and characterization of a specific receptor for mouse oncostatin M.Mol Cell Biol. 1998 Jun;18(6):3357-67. doi: 10.1128/MCB.18.6.3357. Mol Cell Biol. 1998. PMID: 9584176 Free PMC article.
-
Interleukin-6 Family of Cytokines in Cancers.J Interferon Cytokine Res. 2024 Feb;44(2):45-59. doi: 10.1089/jir.2023.0103. Epub 2024 Jan 17. J Interferon Cytokine Res. 2024. PMID: 38232478 Review.
-
[Function, molecular structure and gene expression regulation of receptor for D-factor/LIF].Nihon Rinsho. 1992 Aug;50(8):1956-61. Nihon Rinsho. 1992. PMID: 1433987 Review. Japanese.
Cited by
-
Elastin-like Recombinamers as a Biotechnological Platform for the Development of Cytokine-Functionalized Materials.ACS Omega. 2024 Nov 12;9(47):46733-46742. doi: 10.1021/acsomega.3c09325. eCollection 2024 Nov 26. ACS Omega. 2024. PMID: 39619521 Free PMC article.
References
-
- Hirano T, Yasukawa K, Harada H, et al. Complementary DNA for a novel human interleukin (BSF‐2) that induces B lymphocytes to produce immunoglobulin. Nature. 1986;324:73–76. - PubMed
-
- Murakami M, Kamimura D, Hirano T. Pleiotropy and specificity: Insights from the interleukin 6 family of cytokines. Immunity. 2019;50:812–831. - PubMed
-
- Pinho V, Fernandes M, da Costa A, Machado R, Gomes AC. Leukemia inhibitory factor: Recent advances and implications in biotechnology. Cytokine Growth Factor Rev. 2020;52:25–33. - PubMed
-
- Kazakov AS, Sokolov AS, Permyakova ME, et al. Specific cytokines of interleukin‐6 family interact with S100 proteins. Cell Calcium. 2022;101:102520. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

