Tumor elimination by clustered microRNAs miR-306 and miR-79 via noncanonical activation of JNK signaling
- PMID: 36222503
- PMCID: PMC9612915
- DOI: 10.7554/eLife.77340
Tumor elimination by clustered microRNAs miR-306 and miR-79 via noncanonical activation of JNK signaling
Abstract
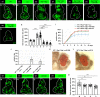
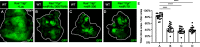
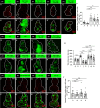
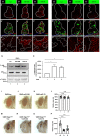
JNK signaling plays a critical role in both tumor promotion and tumor suppression. Here, we identified clustered microRNAs (miRNAs) miR-306 and miR-79 as novel tumor-suppressor miRNAs that specifically eliminate JNK-activated tumors in Drosophila. While showing only a slight effect on normal tissue growth, miR-306 and miR-79 strongly suppressed growth of multiple tumor models, including malignant tumors caused by Ras activation and cell polarity defects. Mechanistically, these miRNAs commonly target the mRNA of an E3 ubiquitin ligase ring finger protein 146 (RNF146). We found that RNF146 promotes degradation of tankyrase (Tnks), an ADP-ribose polymerase that promotes JNK activation in a noncanonical manner. Thus, downregulation of RNF146 by miR-306 and miR-79 leads to hyper-enhancement of JNK activation. Our data show that, while JNK activity is essential for tumor growth, elevation of miR-306 or miR-79 overactivate JNK signaling to the lethal level via noncanonical JNK pathway and thus eliminate tumors, providing a new miRNA-based strategy against cancer.
Keywords: D. melanogaster; JNK signaling; cell biology; cell death; microRNA; tumor suppression.
© 2022, Wang et al.
Conflict of interest statement
ZW, XX, JL, TI No competing interests declared
Figures




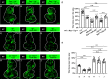





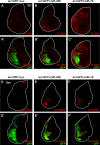
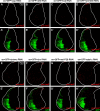

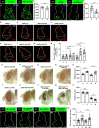


Similar articles
-
Overexpressed microRNA-136 works as a cancer suppressor in gallbladder cancer through suppression of JNK signaling pathway via inhibition of MAP2K4.Am J Physiol Gastrointest Liver Physiol. 2019 Nov 1;317(5):G670-G681. doi: 10.1152/ajpgi.00055.2019. Epub 2019 Aug 1. Am J Physiol Gastrointest Liver Physiol. 2019. PMID: 31369289
-
MicroRNA-370 functions as a tumor suppressor in hepatocellular carcinoma via inhibition of the MAPK/JNK signaling pathway by targeting BEX2.J Hum Genet. 2019 Dec;64(12):1203-1217. doi: 10.1038/s10038-019-0653-x. Epub 2019 Sep 17. J Hum Genet. 2019. PMID: 31530937
-
Structural basis for tankyrase-RNF146 interaction reveals noncanonical tankyrase-binding motifs.Protein Sci. 2018 Jun;27(6):1057-1067. doi: 10.1002/pro.3413. Epub 2018 Apr 25. Protein Sci. 2018. PMID: 29604130 Free PMC article.
-
Regulation of the MIR155 host gene in physiological and pathological processes.Gene. 2013 Dec 10;532(1):1-12. doi: 10.1016/j.gene.2012.12.009. Epub 2012 Dec 14. Gene. 2013. PMID: 23246696 Review.
-
Ring finger protein 146/Iduna is a poly(ADP-ribose) polymer binding and PARsylation dependent E3 ubiquitin ligase.Cell Adh Migr. 2011 Nov-Dec;5(6):463-71. doi: 10.4161/cam.5.6.18356. Cell Adh Migr. 2011. PMID: 22274711 Free PMC article. Review.
Cited by
-
Identification and Characterization of Development-Related microRNAs in the Red Flour Beetle, Tribolium castaneum.Int J Mol Sci. 2023 Apr 3;24(7):6685. doi: 10.3390/ijms24076685. Int J Mol Sci. 2023. PMID: 37047657 Free PMC article.
-
Targeting the PARylation-Dependent Ubiquitination Signaling Pathway for Cancer Therapies.Biomolecules. 2025 Feb 7;15(2):237. doi: 10.3390/biom15020237. Biomolecules. 2025. PMID: 40001540 Free PMC article. Review.
-
Computational Screening to Predict MicroRNA Targets in the Flavivirus 3' UTR Genome: An Approach for Antiviral Development.Int J Mol Sci. 2024 Sep 21;25(18):10135. doi: 10.3390/ijms251810135. Int J Mol Sci. 2024. PMID: 39337625 Free PMC article.
-
Effects of miR-306 Perturbation on Life Parameters in the English Grain Aphid, Sitobion avenae (Homoptera: Aphididae).Int J Mol Sci. 2024 May 23;25(11):5680. doi: 10.3390/ijms25115680. Int J Mol Sci. 2024. PMID: 38891867 Free PMC article.
-
Blockade of Crk eliminates Yki/YAP-activated tumors via JNK-mediated apoptosis in Drosophila.Commun Biol. 2024 Sep 28;7(1):1196. doi: 10.1038/s42003-024-06897-w. Commun Biol. 2024. PMID: 39341909 Free PMC article.
References
-
- Adachi-Yamada T, Gotoh T, Sugimura I, Tateno M, Nishida Y, Onuki T, Date H. De novo synthesis of sphingolipids is required for cell survival by down-regulating c-Jun N-terminal kinase in Drosophila imaginal discs. Molecular and Cellular Biology. 1999;19:7276–7286. doi: 10.1128/MCB.19.10.7276. - DOI - PMC - PubMed
-
- Beg MS, Brenner AJ, Sachdev J, Borad M, Kang YK, Stoudemire J, Smith S, Bader AG, Kim S, Hong DS. Phase I study of MRX34, a liposomal miR-34a mimic, administered twice Weekly in patients with advanced solid tumors. Investigational New Drugs. 2017;35:180–188. doi: 10.1007/s10637-016-0407-y. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

