Leveraging a pharmacogenomics knowledgebase to formulate a drug response phenotype terminology for genomic medicine
- PMID: 36222570
- PMCID: PMC9710557
- DOI: 10.1093/bioinformatics/btac646
Leveraging a pharmacogenomics knowledgebase to formulate a drug response phenotype terminology for genomic medicine
Abstract
Motivation: Despite the increasing evidence of utility of genomic medicine in clinical practice, systematically integrating genomic medicine information and knowledge into clinical systems with a high-level of consistency, scalability and computability remains challenging. A comprehensive terminology is required for relevant concepts and the associated knowledge model for representing relationships. In this study, we leveraged PharmGKB, a comprehensive pharmacogenomics (PGx) knowledgebase, to formulate a terminology for drug response phenotypes that can represent relationships between genetic variants and treatments. We evaluated coverage of the terminology through manual review of a randomly selected subset of 200 sentences extracted from genetic reports that contained concepts for 'Genes and Gene Products' and 'Treatments'.
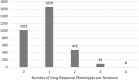
Results: Results showed that our proposed drug response phenotype terminology could cover 96% of the drug response phenotypes in genetic reports. Among 18 653 sentences that contained both 'Genes and Gene Products' and 'Treatments', 3011 sentences were able to be mapped to a drug response phenotype in our proposed terminology, among which the most discussed drug response phenotypes were response (994), sensitivity (829) and survival (332). In addition, we were able to re-analyze genetic report context incorporating the proposed terminology and enrich our previously proposed PGx knowledge model to reveal relationships between genetic variants and treatments. In conclusion, we proposed a drug response phenotype terminology that enhanced structured knowledge representation of genomic medicine.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2022. Published by Oxford University Press. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures

Similar articles
-
Simplifying the use of pharmacogenomics in clinical practice: Building the genomic prescribing system.J Biomed Inform. 2017 Nov;75:110-121. doi: 10.1016/j.jbi.2017.09.012. Epub 2017 Sep 28. J Biomed Inform. 2017. PMID: 28963061
-
Pharmacogenomics knowledge for personalized medicine.Clin Pharmacol Ther. 2012 Oct;92(4):414-7. doi: 10.1038/clpt.2012.96. Clin Pharmacol Ther. 2012. PMID: 22992668 Free PMC article. Review.
-
PharmGKB Tutorial for Pharmacogenomics of Drugs Potentially Used in the Context of COVID-19.Clin Pharmacol Ther. 2021 Jan;109(1):116-122. doi: 10.1002/cpt.2067. Epub 2020 Oct 24. Clin Pharmacol Ther. 2021. PMID: 32978778 Free PMC article.
-
PharmGKB, an Integrated Resource of Pharmacogenomic Knowledge.Curr Protoc. 2021 Aug;1(8):e226. doi: 10.1002/cpz1.226. Curr Protoc. 2021. PMID: 34387941 Free PMC article.
-
Genomic medicine in Africa: a need for molecular genetics and pharmacogenomics experts.Curr Med Res Opin. 2023 Jan;39(1):141-147. doi: 10.1080/03007995.2022.2124072. Epub 2022 Sep 19. Curr Med Res Opin. 2023. PMID: 36094413 Review.
References
-
- Antoniou G., Van Harmelen F. (2004) Web ontology language: OWL. In: Staab,S. and Studer,R. (eds.) Handbook on Ontologies. Springer. pp. 67–92.
-
- Aronson A.R. (2006) Metamap: Mapping Text to the UMLS Metathesaurus. NLM, NIH, DHHS, Bethesda, MD, pp. 1–26.

