ZAXINONE SYNTHASE 2 regulates growth and arbuscular mycorrhizal symbiosis in rice
- PMID: 36222582
- PMCID: PMC9806602
- DOI: 10.1093/plphys/kiac472
ZAXINONE SYNTHASE 2 regulates growth and arbuscular mycorrhizal symbiosis in rice
Abstract
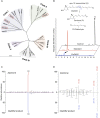
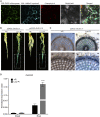
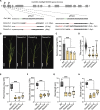
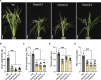
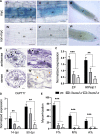
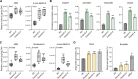
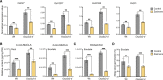
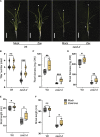
Carotenoid cleavage, catalyzed by CAROTENOID CLEAVAGE DIOXYGENASEs (CCDs), provides signaling molecules and precursors of plant hormones. Recently, we showed that zaxinone, a apocarotenoid metabolite formed by the CCD ZAXINONE SYNTHASE (ZAS), is a growth regulator required for normal rice (Oryza sativa) growth and development. The rice genome encodes three OsZAS homologs, called here OsZAS1b, OsZAS1c, and OsZAS2, with unknown functions. Here, we investigated the enzymatic activity, expression pattern, and subcellular localization of OsZAS2 and generated and characterized loss-of-function CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats and associated protein 9)-Oszas2 mutants. We show that OsZAS2 formed zaxinone in vitro. OsZAS2 was predominantly localized in plastids and mainly expressed under phosphate starvation. Moreover, OsZAS2 expression increased during mycorrhization, specifically in arbuscule-containing cells. Oszas2 mutants contained lower zaxinone content in roots and exhibited reduced root and shoot biomass, fewer tillers, and higher strigolactone (SL) levels. Exogenous zaxinone application repressed SL biosynthesis and partially rescued the growth retardation of the Oszas2 mutant. Consistent with the OsZAS2 expression pattern, Oszas2 mutants displayed a lower frequency of arbuscular mycorrhizal colonization. In conclusion, OsZAS2 is a zaxinone-forming enzyme that, similar to the previously reported OsZAS, determines rice growth, architecture, and SL content, and is required for optimal mycorrhization.
© The Author(s) 2022. Published by Oxford University Press on behalf of American Society of Plant Biologists.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

