Resolving subcellular pH with a quantitative fluorescent lifetime biosensor
- PMID: 36224168
- PMCID: PMC9556823
- DOI: 10.1038/s41467-022-33348-z
Resolving subcellular pH with a quantitative fluorescent lifetime biosensor
Abstract
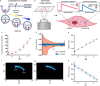
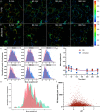
Changes in sub-cellular pH play a key role in metabolism, membrane transport, and triggering cargo release from therapeutic delivery systems. Most methods to measure pH rely on intensity changes of pH sensitive fluorophores, however, these measurements are hampered by high uncertainty in the inferred pH and the need for multiple fluorophores. To address this, here we combine pH dependant fluorescent lifetime imaging microscopy (pHLIM) with deep learning to accurately quantify sub-cellular pH in individual vesicles. We engineer the pH sensitive protein mApple to localise in the cytosol, endosomes, and lysosomes, and demonstrate that pHLIM can rapidly detect pH changes induced by drugs such as bafilomycin A1 and chloroquine. We also demonstrate that polyethylenimine (a common transfection reagent) does not exhibit a proton sponge effect and had no measurable impact on the pH of endocytic vesicles. pHLIM is a simple and quantitative method that will help to understand drug action and disease progression.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Research Materials

