Clinical phenotypes and outcomes associated with SARS-CoV-2 variant Omicron in critically ill French patients with COVID-19
- PMID: 36224216
- PMCID: PMC9555693
- DOI: 10.1038/s41467-022-33801-z
Clinical phenotypes and outcomes associated with SARS-CoV-2 variant Omicron in critically ill French patients with COVID-19
Erratum in
-
Author Correction: Clinical phenotypes and outcomes associated with SARS-CoV-2 variant Omicron in critically ill French patients with COVID-19.Nat Commun. 2022 Dec 7;13(1):7547. doi: 10.1038/s41467-022-34575-0. Nat Commun. 2022. PMID: 36477670 Free PMC article. No abstract available.
Abstract
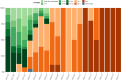
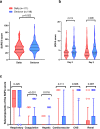
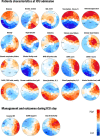
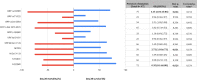
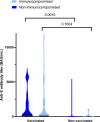
Infection with SARS-CoV-2 variant Omicron is considered to be less severe than infection with variant Delta, with rarer occurrence of severe disease requiring intensive care. Little information is available on comorbid factors, clinical conditions and specific viral mutational patterns associated with the severity of variant Omicron infection. In this multicenter prospective cohort study, patients consecutively admitted for severe COVID-19 in 20 intensive care units in France between December 7th 2021 and May 1st 2022 were included. Among 259 patients, we show that the clinical phenotype of patients infected with variant Omicron (n = 148) is different from that in those infected with variant Delta (n = 111). We observe no significant relationship between Delta and Omicron variant lineages/sublineages and 28-day mortality (adjusted odds ratio [95% confidence interval] = 0.68 [0.35-1.32]; p = 0.253). Among Omicron-infected patients, 43.2% are immunocompromised, most of whom have received two doses of vaccine or more (85.9%) but display a poor humoral response to vaccination. The mortality rate of immunocompromised patients infected with variant Omicron is significantly higher than that of non-immunocompromised patients (46.9% vs 26.2%; p = 0.009). In patients infected with variant Omicron, there is no association between specific sublineages (BA.1/BA.1.1 (n = 109) and BA.2 (n = 21)) or any viral genome polymorphisms/mutational profile and 28-day mortality.
© 2022. The Author(s).
Conflict of interest statement
S.F. has served as a speaker for GlaxoSmithKline, Abbvie, and Abbott Diagnostics. J.-M.P. has served as an advisor or speaker for Abbvie, Arbutus, Assembly Biosciences, Gilead and Merck. E.A. has received fees for lectures from Alexion, Sanofi, Gilead and Pfizer. His hospital has received research grant from Pfizer, MSD and Alexion. D.D. served as an advisor for Gilead-Sciences, ViiV Health care, Janssen-Cilag et MSD. F.P. served as an advisor for Gilead; he also received research grant from Alexion. C.-E.L. received lecture fees from MSD, Aerogen, Advanzpharma, and BioMérieux, outside the submitted work. J.-F.T. served as an advisor for pfizer, Gilead, BD, Gilead, Merck; he also received research grant from Thermofischer, merck, Pfizer, Biomerieux; lectures: pfizer, biomerieux BD, Merck, Shionoghi outside the submitted work. Other authors have no conflict of interest to disclose.
Figures
References
-
- Menni, C. et al. Symptom prevalence, duration, and risk of hospital admission in individuals infected with SARS-CoV-2 during periods of omicron and delta variant dominance: a prospective observational study from the ZOE COVID Study. Lancet S0140673622003270. 10.1016/S0140-6736(22)00327-0 (2022). - PMC - PubMed
Publication types
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

