Universal dynamics of mitochondrial networks: a finite-size scaling analysis
- PMID: 36224243
- PMCID: PMC9556628
- DOI: 10.1038/s41598-022-14946-9
Universal dynamics of mitochondrial networks: a finite-size scaling analysis
Abstract
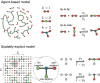
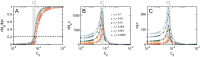
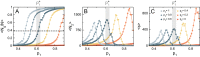
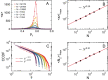
Evidence from models and experiments suggests that the networked structure observed in mitochondria emerges at the critical point of a phase transition controlled by fission and fusion rates. If mitochondria are poised at criticality, the relevant network quantities should scale with the system's size. However, whether or not the expected finite-size effects take place has not been demonstrated yet. Here, we first provide a theoretical framework to interpret the scaling behavior of mitochondrial network quantities by analyzing two conceptually different models of mitochondrial dynamics. Then, we perform a finite-size scaling analysis of real mitochondrial networks extracted from microscopy images and obtain scaling exponents comparable with critical exponents from models and theory. Overall, we provide a universal description of the structural phase transition in mammalian mitochondria.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Mitochondrial network complexity emerges from fission/fusion dynamics.Sci Rep. 2018 Jan 10;8(1):363. doi: 10.1038/s41598-017-18351-5. Sci Rep. 2018. PMID: 29321534 Free PMC article.
-
Finite-size scaling of survival probability in branching processes.Phys Rev E Stat Nonlin Soft Matter Phys. 2015 Apr;91(4):042122. doi: 10.1103/PhysRevE.91.042122. Epub 2015 Apr 20. Phys Rev E Stat Nonlin Soft Matter Phys. 2015. PMID: 25974453
-
Universal organization of resting brain activity at the thermodynamic critical point.Front Syst Neurosci. 2013 Aug 22;7:42. doi: 10.3389/fnsys.2013.00042. eCollection 2013. Front Syst Neurosci. 2013. PMID: 23986660 Free PMC article.
-
Mitochondrial Function and Cell Size: An Allometric Relationship.Trends Cell Biol. 2017 Jun;27(6):393-402. doi: 10.1016/j.tcb.2017.02.006. Epub 2017 Mar 8. Trends Cell Biol. 2017. PMID: 28284466 Review.
-
Mechanisms of mitochondrial reorganization.J Biochem. 2024 Feb 25;175(2):167-178. doi: 10.1093/jb/mvad098. J Biochem. 2024. PMID: 38016932 Review.
Cited by
-
Finite-Size Scaling in the Ageing Dynamics of the 1D Glauber-Ising Model.Entropy (Basel). 2025 Jan 28;27(2):139. doi: 10.3390/e27020139. Entropy (Basel). 2025. PMID: 40003136 Free PMC article.
-
Mitochondrial networks through the lens of mathematics.Phys Biol. 2023 Jul 14;20(5):051001. doi: 10.1088/1478-3975/acdcdb. Phys Biol. 2023. PMID: 37290456 Free PMC article. Review.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

