A comparative study of spike protein of SARS-CoV-2 and its variant Omicron (B.1.1.529) on some immune characteristics
- PMID: 36224298
- PMCID: PMC9554390
- DOI: 10.1038/s41598-022-21690-7
A comparative study of spike protein of SARS-CoV-2 and its variant Omicron (B.1.1.529) on some immune characteristics
Abstract
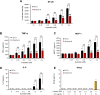
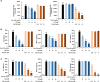
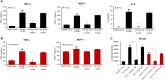
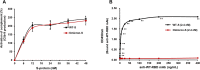
The emergence of Omicron variant raises great concerns because of its rapid transmissibility and its numerous mutations in spike protein (S-protein). S-protein can act as a pathogen-associated molecular pattern and complement activator as well as antigen. We compared some immune characteristics of trimer S-proteins for wild type (WT-S) and B.1.1.529 Omicron (Omicron-S) to investigate whether the mutations have affected its pathogenicity and antigenic shift. The results indicated that WT-S and Omicron-S directly activated nuclear factor-κB (NF-κB) and induced the release of pro-inflammatory cytokines in macrophages, but the actions of Omicron-S were weaker. These inflammatory reactions could be abrogated by a Toll-like receptor 4 antagonist TAK-242. Two S-proteins failed to induce the production of antiviral molecular interferon-β. In contrast to pro-inflammatory effects, the ability of two S-proteins to activate complement was comparable. We also compared the binding ability of two S-proteins to a high-titer anti-WT-receptor-binding domain antibody. The data showed that WT-S strongly bound to this antibody, while Omicron-S was completely off-target. Collectively, the mutations of Omicron have a great impact on the pro-inflammatory ability and epitopes of S-protein, but little effect on its ability to activate complement. Addressing these issues can be helpful for more adequate understanding of the pathogenicity of Omicron and the vaccine breakthrough infection.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

