Decoding the transcriptome of calcified atherosclerotic plaque at single-cell resolution
- PMID: 36224302
- PMCID: PMC9556750
- DOI: 10.1038/s42003-022-04056-7
Decoding the transcriptome of calcified atherosclerotic plaque at single-cell resolution
Abstract
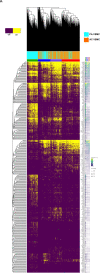
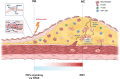
Atherogenesis involves an interplay of inflammation, tissue remodeling and cellular transdifferentiation (CTD), making it especially difficult to precisely delineate its pathophysiology. Here we use single-cell RNA sequencing and systems-biology approaches to analyze the transcriptional profiles of vascular smooth muscle cells (VSMCs) and endothelial cells (ECs) in calcified atherosclerotic core (AC) plaques and patient-matched proximal adjacent (PA) portions of carotid artery tissue from patients undergoing carotid endarterectomy. Our results reveal an anatomic distinction whereby PA cells express inflammatory mediators, while cells expressing matrix-secreting genes occupy a majority of the AC region. Systems biology analysis indicates that inflammation in PA ECs and VSMCs may be driven by TNFa signaling. Furthermore, we identify POSTN, SPP1 and IBSP in AC VSMCs, and ITLN1, SCX and S100A4 in AC ECs as possible candidate drivers of CTD in the atherosclerotic core. These results establish an anatomic framework for atherogenesis which forms the basis for exploration of a site-specific strategy for disruption of disease progression.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







References
-
- Basatemur, G. L., Jørgensen, H. F., Clarke, M. C. H., Bennett, M. R. & Mallat, Z. Vascular smooth muscle cells in atherosclerosis. Nat. Rev. Cardiol.10.1038/s41569-019-0227-9 (2019). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

