Activation of γ-globin expression by hypoxia-inducible factor 1α
- PMID: 36224385
- PMCID: PMC9773321
- DOI: 10.1038/s41586-022-05312-w
Activation of γ-globin expression by hypoxia-inducible factor 1α
Abstract
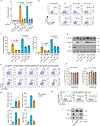
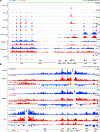
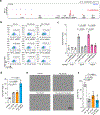
Around birth, globin expression in human red blood cells (RBCs) shifts from γ-globin to β-globin, which results in fetal haemoglobin (HbF, α2γ2) being gradually replaced by adult haemoglobin (HbA, α2β2)1. This process has motivated the development of innovative approaches to treat sickle cell disease and β-thalassaemia by increasing HbF levels in postnatal RBCs2. Here we provide therapeutically relevant insights into globin gene switching obtained through a CRISPR-Cas9 screen for ubiquitin-proteasome components that regulate HbF expression. In RBC precursors, depletion of the von Hippel-Lindau (VHL) E3 ubiquitin ligase stabilized its ubiquitination target, hypoxia-inducible factor 1α (HIF1α)3,4, to induce γ-globin gene transcription. Mechanistically, HIF1α-HIF1β heterodimers bound cognate DNA elements in BGLT3, a long noncoding RNA gene located 2.7 kb downstream of the tandem γ-globin genes HBG1 and HBG2. This was followed by the recruitment of transcriptional activators, chromatin opening and increased long-range interactions between the γ-globin genes and their upstream enhancer. Similar induction of HbF occurred with hypoxia or with inhibition of prolyl hydroxylase domain enzymes that target HIF1α for ubiquitination by the VHL E3 ubiquitin ligase. Our findings link globin gene regulation with canonical hypoxia adaptation, provide a mechanism for HbF induction during stress erythropoiesis and suggest a new therapeutic approach for β-haemoglobinopathies.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests
M.J.W. serves on the advisory boards for Cellarity Inc., Novartis, Graphite Bio, Dyne Therapeutics, and Forma Therapeutics, and owns equity in Cellarity, Inc.
Figures







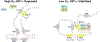


Comment in
-
HIF1α reboots fetal haemoglobin production.Nat Rev Drug Discov. 2022 Dec;21(12):878. doi: 10.1038/d41573-022-00179-y. Nat Rev Drug Discov. 2022. PMID: 36307651 No abstract available.
References
Main Text References
-
- Vinjamur DS, Bauer DE & Orkin SH Recent progress in understanding and manipulating haemoglobin switching for the haemoglobinopathies. Br J Haematol 180, 630–643 (2018). - PubMed
-
- Kaelin WG Jr. & Ratcliffe PJ Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell 30, 393–402 (2008). - PubMed
-
- Semenza GL Hypoxia-inducible factor 1 (HIF-1) pathway. Sci STKE 2007, cm8 (2007). - PubMed
-
- Kato GJ, et al. Sickle cell disease. Nat Rev Dis Primers 4, 18010 (2018). - PubMed
References for Methods
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

