The FUS/circEZH2/KLF5/ feedback loop contributes to CXCR4-induced liver metastasis of breast cancer by enhancing epithelial-mesenchymal transition
- PMID: 36224562
- PMCID: PMC9555172
- DOI: 10.1186/s12943-022-01653-2
The FUS/circEZH2/KLF5/ feedback loop contributes to CXCR4-induced liver metastasis of breast cancer by enhancing epithelial-mesenchymal transition
Abstract
Background: Metastasis of breast cancer have caused the majority of cancer-related death worldwide. The circRNAs are associated with tumorigenesis and metastasis in breast cancer according to recent research. However, the biological mechanism of circRNAs in liver metastatic breast cancer remains ambiguous yet.
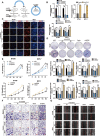
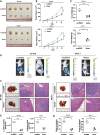
Methods: Microarray analysis of three pairs of primary BC tissues and matched hepatic metastatic specimens identified circEZH2. We used RT-qPCR and FISH assays to confirm circEZH2 existence, characteristics, and expression. Both in vivo and in vitro, circEZH2 played an oncogenic role which promoted metastasis as well. A range of bioinformatic analysis, Western blot, RNA pull-down, RIP, ChIP, and animal experiments were used to define the feedback loop involving FUS, circEZH2, miR-217-5p, KLF5, FUS, CXCR4 as well as epithelial and mesenchymal transition.
Results: In our research, circEZH2 was proved to be upregulated in liver metastases in BC and predicted the worse prognosis in breast cancer patients. Overexpression of circEZH2 notably accentuated the vitality and invasion of BC cells, whereas knockdown of circEZH2 elicited the literally opposite effects. Besides, overexpressed circEZH2 promoted tumorigenesis and liver metastasis in vivo. Moreover, circEZH2 could adsorb miR-217-5p to upregulate KLF5 thus leading to activate FUS transcription which would facilitate the back-splicing program of circEZH2. Meanwhile, KLF5 could upregulated CXCR4 transcriptionally to accelerate epithelial and mesenchymal transition of breast cancer.
Conclusions: Consequently, a novel feedback loop FUS/circEZH2/KLF5/CXCR4 was established while circEZH2 could be novel biomarker and potential target for BC patients' therapy.
Keywords: Breast cancer; EMT; Feedback loop; Metastasis; circRNAs.
© 2022. The Author(s).
Conflict of interest statement
The authors have disclosed no competing interests.
Figures






Similar articles
-
The circROBO1/KLF5/FUS feedback loop regulates the liver metastasis of breast cancer by inhibiting the selective autophagy of afadin.Mol Cancer. 2022 Jan 24;21(1):29. doi: 10.1186/s12943-022-01498-9. Mol Cancer. 2022. PMID: 35073911 Free PMC article.
-
CEP55 3'-UTR promotes epithelial-mesenchymal transition and enhances tumorigenicity of bladder cancer cells by acting as a ceRNA regulating miR-497-5p.Cell Oncol (Dordr). 2022 Dec;45(6):1217-1236. doi: 10.1007/s13402-022-00712-6. Epub 2022 Nov 14. Cell Oncol (Dordr). 2022. PMID: 36374443
-
Lidocaine inhibits glioma cell proliferation, migration and invasion by modulating the circEZH2/miR-181b-5p pathway.Neuroreport. 2021 Jan 6;32(1):52-60. doi: 10.1097/WNR.0000000000001560. Neuroreport. 2021. PMID: 33252475
-
CircGPR137B/miR-4739/FTO feedback loop suppresses tumorigenesis and metastasis of hepatocellular carcinoma.Mol Cancer. 2022 Jul 20;21(1):149. doi: 10.1186/s12943-022-01619-4. Mol Cancer. 2022. PMID: 35858900 Free PMC article.
-
Tumor-on-chip platforms for breast cancer continuum concept modeling.Front Bioeng Biotechnol. 2024 Oct 2;12:1436393. doi: 10.3389/fbioe.2024.1436393. eCollection 2024. Front Bioeng Biotechnol. 2024. PMID: 39416279 Free PMC article. Review.
Cited by
-
Intermediate Filaments in Breast Cancer Progression, and Potential Biomarker for Cancer Therapy: A Narrative Review.Breast Cancer (Dove Med Press). 2024 Oct 14;16:689-704. doi: 10.2147/BCTT.S489953. eCollection 2024. Breast Cancer (Dove Med Press). 2024. PMID: 39430570 Free PMC article. Review.
-
The role and application of vesicles in triple-negative breast cancer: Opportunities and challenges.Mol Ther Oncolytics. 2023 Nov 23;31:100752. doi: 10.1016/j.omto.2023.100752. eCollection 2023 Dec 19. Mol Ther Oncolytics. 2023. PMID: 38130701 Free PMC article. Review.
-
Targeting tumor exosomal circular RNA cSERPINE2 suppresses breast cancer progression by modulating MALT1-NF-𝜅B-IL-6 axis of tumor-associated macrophages.J Exp Clin Cancer Res. 2023 Feb 17;42(1):48. doi: 10.1186/s13046-023-02620-5. J Exp Clin Cancer Res. 2023. PMID: 36797769 Free PMC article.
-
circMMD reduction following tumor treating fields inhibits glioblastoma progression through FUBP1/FIR/DVL1 and miR-15b-5p/FZD6 signaling.J Exp Clin Cancer Res. 2023 Mar 17;42(1):64. doi: 10.1186/s13046-023-02642-z. J Exp Clin Cancer Res. 2023. PMID: 36932454 Free PMC article.
-
circROBO1 promotes prostate cancer growth and enzalutamide resistance via accelerating glycolysis.J Cancer. 2023 Aug 21;14(13):2574-2584. doi: 10.7150/jca.86940. eCollection 2023. J Cancer. 2023. PMID: 37670963 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

