Detection of SARS-CoV-2 in a dog with hemorrhagic diarrhea
- PMID: 36224622
- PMCID: PMC9554378
- DOI: 10.1186/s12917-022-03453-8
Detection of SARS-CoV-2 in a dog with hemorrhagic diarrhea
Abstract
Background: Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of COVID-19, has infected several animal species, including dogs, presumably via human-to-animal transmission. Most infected dogs reported were asymptomatic, with low viral loads. However, in this case we detected SARS-CoV-2 in a dog from the North African coastal Spanish city of Ceuta presenting hemorrhagic diarrhea, a disease also reported earlier on in an infected dog from the USA.
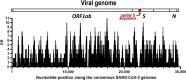
Case presentation: In early January 2021, a West Highland Terrier pet dog from Ceuta (Spain) presented hemorrhagic diarrhea with negative tests for candidate microbial pathogens. Since the animal was in a household whose members suffered SARS-CoV-2 in December 2020, dog feces were analyzed for SARS-CoV-2, proving positive in a two-tube RT-PCR test, with confirmation by sequencing a 399-nucleotide region of the spike (S) gene. Furthermore, next-generation sequencing (NGS) covered > 90% SARS-CoV-2 genome sequence, allowing to classify it as variant B.1.177. Remarkably, the sequence revealed the Ile402Val substitution in the spike protein (S), of potential concern because it mapped in the receptor binding domain (RBD) that mediates virus interaction with the cell. NGS reads mapping to bacterial genomes showed that the dog fecal microbiome fitted best the characteristic microbiome of dog's acute hemorrhagic diarrhea.
Conclusion: Our findings exemplify dog infection stemming from the human SARS-CoV-2 pandemic, providing nearly complete-genome sequencing of the virus, which is recognized as belonging to the B.1.177 variant, adding knowledge on variant circulation in a geographic region and period for which there was little viral variant characterization. A single amino acid substitution found in the S protein that could have been of concern is excluded to belong to this category given its rarity and intrinsic nature. The dog's pathology suggests that SARS-CoV-2 could affect the gastrointestinal tract of the dog.
Keywords: B.1.177; Dog COVID-19; Ile402Val S protein substitution; One Health; SARS-CoV-2; Zoonosis.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
First Detection of SARS-CoV-2 B.1.1.7 Variant of Concern in an Asymptomatic Dog in Spain.Viruses. 2021 Jul 15;13(7):1379. doi: 10.3390/v13071379. Viruses. 2021. PMID: 34372585 Free PMC article.
-
First Detection of SARS-CoV-2 Delta (B.1.617.2) Variant of Concern in a Dog with Clinical Signs in Spain.Viruses. 2021 Dec 16;13(12):2526. doi: 10.3390/v13122526. Viruses. 2021. PMID: 34960795 Free PMC article.
-
First evidence of human-to-dog transmission of SARS-CoV-2 B.1.160 variant in France.Transbound Emerg Dis. 2022 Jul;69(4):e823-e830. doi: 10.1111/tbed.14359. Epub 2021 Nov 8. Transbound Emerg Dis. 2022. PMID: 34706153 Free PMC article.
-
SARS-CoV-2's Variants of Concern: A Brief Characterization.Front Immunol. 2022 Jul 26;13:834098. doi: 10.3389/fimmu.2022.834098. eCollection 2022. Front Immunol. 2022. PMID: 35958548 Free PMC article. Review.
-
Transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) to animals: an updated review.J Transl Med. 2020 Sep 21;18(1):358. doi: 10.1186/s12967-020-02534-2. J Transl Med. 2020. PMID: 32957995 Free PMC article. Review.
Cited by
-
A SARS-CoV-2 full genome sequence of the B.1.1 lineage sheds light on viral evolution in Sicily in late 2020.Front Public Health. 2023 Jan 26;11:1098965. doi: 10.3389/fpubh.2023.1098965. eCollection 2023. Front Public Health. 2023. PMID: 36778569 Free PMC article.
-
Serological screening of SARS-CoV-2 infection in companion animals of Buenos Aires suburbs.Front Vet Sci. 2023 May 30;10:1161820. doi: 10.3389/fvets.2023.1161820. eCollection 2023. Front Vet Sci. 2023. PMID: 37323839 Free PMC article.
-
SARS-CoV-2 in Namibian Dogs.Vaccines (Basel). 2022 Dec 13;10(12):2134. doi: 10.3390/vaccines10122134. Vaccines (Basel). 2022. PMID: 36560544 Free PMC article.
-
Development and Validation of a Panel of One-Step Four-Plex qPCR/RT-qPCR Assays for Simultaneous Detection of SARS-CoV-2 and Other Pathogens Associated with Canine Infectious Respiratory Disease Complex.Viruses. 2023 Sep 5;15(9):1881. doi: 10.3390/v15091881. Viruses. 2023. PMID: 37766287 Free PMC article.
-
SARS-CoV-2 exposure in hunting and stray dogs of southern Italy.Vet Res Commun. 2024 Dec;48(6):4037-4042. doi: 10.1007/s11259-024-10496-9. Epub 2024 Aug 21. Vet Res Commun. 2024. PMID: 39167256 Free PMC article.
References
-
- OIE, 2022. https://www.oie.int/en/what-we-offer/emergency-and-resilience/covid-19/#.... Accessed on 17 May 2022.
-
- Lopes Melo F, Bezerra B, Oliveira Luna F, Navarrete Barragan NA, Loyo Arcoverde RM, Umeed R, et al. Coronavirus (SARS-CoV-2) in Antillean manatees (Trichechus manatus manatus). Res Square. 2022. 10.21203/rs.3.rs-1065379/v1.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

