Development versus predation: Transcriptomic changes during the lifecycle of Myxococcus xanthus
- PMID: 36225384
- PMCID: PMC9548883
- DOI: 10.3389/fmicb.2022.1004476
Development versus predation: Transcriptomic changes during the lifecycle of Myxococcus xanthus
Abstract
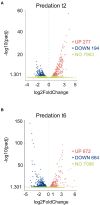
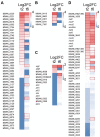
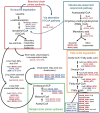
Myxococcus xanthus is a multicellular bacterium with a complex lifecycle. It is a soil-dwelling predator that preys on a wide variety of microorganisms by using a group and collaborative epibiotic strategy. In the absence of nutrients this myxobacterium enters in a unique developmental program by using sophisticated and complex regulatory systems where more than 1,400 genes are transcriptional regulated to guide the community to aggregate into macroscopic fruiting bodies filled of environmentally resistant myxospores. Herein, we analyze the predatosome of M. xanthus, that is, the transcriptomic changes that the predator undergoes when encounters a prey. This study has been carried out using as a prey Sinorhizobium meliloti, a nitrogen fixing bacteria very important for the fertility of soils. The transcriptional changes include upregulation of genes that help the cells to detect, kill, lyse, and consume the prey, but also downregulation of genes not required for the predatory process. Our results have shown that, as expected, many genes encoding hydrolytic enzymes and enzymes involved in biosynthesis of secondary metabolites increase their expression levels. Moreover, it has been found that the predator modifies its lipid composition and overproduces siderophores to take up iron. Comparison with developmental transcriptome reveals that M. xanthus downregulates the expression of a significant number of genes coding for regulatory elements, many of which have been demonstrated to be key elements during development. This study shows for the first time a global view of the M. xanthus lifecycle from a transcriptome perspective.
Keywords: Myxococcus xanthus; Sinorhizobacterium meliloti; bacterial predation; development; predatosome; transcriptome.
Copyright © 2022 Pérez, Contreras-Moreno, Muñoz-Dorado and Moraleda-Muñoz.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Copper and Melanin Play a Role in Myxococcus xanthus Predation on Sinorhizobium meliloti.Front Microbiol. 2020 Feb 4;11:94. doi: 10.3389/fmicb.2020.00094. eCollection 2020. Front Microbiol. 2020. PMID: 32117124 Free PMC article.
-
Transcriptomic response of Sinorhizobium meliloti to the predatory attack of Myxococcus xanthus.Front Microbiol. 2023 Jun 19;14:1213659. doi: 10.3389/fmicb.2023.1213659. eCollection 2023. Front Microbiol. 2023. PMID: 37405170 Free PMC article.
-
Myxococcus xanthus predation: an updated overview.Front Microbiol. 2024 Jan 24;15:1339696. doi: 10.3389/fmicb.2024.1339696. eCollection 2024. Front Microbiol. 2024. PMID: 38328431 Free PMC article. Review.
-
Dynamics of Solitary Predation by Myxococcus xanthus on Escherichia coli Observed at the Single-Cell Level.Appl Environ Microbiol. 2020 Jan 21;86(3):e02286-19. doi: 10.1128/AEM.02286-19. Print 2020 Jan 21. Appl Environ Microbiol. 2020. PMID: 31704687 Free PMC article.
-
The Predation Strategy of Myxococcus xanthus.Front Microbiol. 2020 Jan 14;11:2. doi: 10.3389/fmicb.2020.00002. eCollection 2020. Front Microbiol. 2020. PMID: 32010119 Free PMC article. Review.
Cited by
-
A Diverged Transcriptional Network for Usage of Two Fe-S Cluster Biogenesis Machineries in the Delta-Proteobacterium Myxococcus xanthus.mBio. 2023 Feb 28;14(1):e0300122. doi: 10.1128/mbio.03001-22. Epub 2023 Jan 19. mBio. 2023. PMID: 36656032 Free PMC article.
-
Two reasons to kill: predation and kin discrimination in myxobacteria.Microbiology (Reading). 2023 Jul;169(7):001372. doi: 10.1099/mic.0.001372. Microbiology (Reading). 2023. PMID: 37494115 Free PMC article. Review.
-
Cell-cell transfer of adaptation traits benefits kin and actor in a cooperative microbe.Proc Natl Acad Sci U S A. 2024 Jul 23;121(30):e2402559121. doi: 10.1073/pnas.2402559121. Epub 2024 Jul 16. Proc Natl Acad Sci U S A. 2024. PMID: 39012831 Free PMC article.
-
A hunting ground for predatory bacteria at the Zhenbei seamount in the South China Sea.ISME Commun. 2025 Mar 5;5(1):ycaf042. doi: 10.1093/ismeco/ycaf042. eCollection 2025 Jan. ISME Commun. 2025. PMID: 40144403 Free PMC article.
-
Comparative genomics and transcriptomics insight into myxobacterial metabolism potentials and multiple predatory strategies.Front Microbiol. 2023 May 5;14:1146523. doi: 10.3389/fmicb.2023.1146523. eCollection 2023. Front Microbiol. 2023. PMID: 37213496 Free PMC article.
References
-
- Altmeyer M. O. (2010). Iron Regulation in the Myxobacterium Myxococcus xanthus DK1622. Master’s thesis, Saarland: University of Saarland.
-
- Arend K. I., Schmidt J. J., Bentler T., Lüchtefeld C., Eggerichs D., Hexamer H. M., et al. . (2021). Myxococcus xanthus predation of gram-positive or gram-negative bacteria is mediated by different bacteriolytic mechanisms. Appl. Environ. Microbiol. 87, e02382–e02320. doi: 10.1128/aem.02382-20 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

