Investigating the association between armour coverage and parasite infection in an estuarine population of stickleback
- PMID: 36226095
- PMCID: PMC9552957
Investigating the association between armour coverage and parasite infection in an estuarine population of stickleback
Abstract
Background: When threespine stickleback colonized fresh water, they repeatedly evolved reduced armour plating via changes in Eda allele frequency. This evolution is typically attributed to differential predation pressure between marine and freshwater environments. However, the chromosomal region containing Eda is associated with many other phenotypes, including schooling, antipredator behaviour, and immunity. Consequently, the evolution of armour plating may be driven by multiple selective pressures acting on Eda or linked genes.
Question: Is parasite infection associated with armour phenotype?
Hypothesis: Parasite load differs between stickleback armour plate morphs.
Organisms: An armour-polymorphic population of threespine stickleback (Gasterosteus aculeatus), and their parasites.
Field site: In June 2009 and 2012, we sampled stickleback from a single human-made salt-marsh pool in the Campbell River Estuary on Vancouver Island.
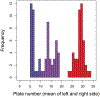
Methods: We counted macroparasites on approximately 100 fish per year and counted lateral armour plates. We used generalized linear models to test for correlations between armour morph and parasite load.
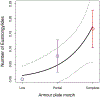
Results: Most parasite species were not associated with armour. The gill parasite Thersitina was more abundant on more fully armoured fish in both years. The nematode Eustrongylides also exhibited a marginally significant positive trend. If parasitic infections reduce stickleback fitness, this positive covariance between armour and infection would accelerate the loss of armour plating in stickleback colonizing fresh water.
Keywords: Ectodysplasin (Eda); Gasterostereus aculeatus; lateral plates; pleiotropy; threespine stickleback.
Figures


Similar articles
-
Adaptation via pleiotropy and linkage: Association mapping reveals a complex genetic architecture within the stickleback Eda locus.Evol Lett. 2020 May 27;4(4):282-301. doi: 10.1002/evl3.175. eCollection 2020 Aug. Evol Lett. 2020. PMID: 32774879 Free PMC article.
-
Ongoing niche differentiation under high gene flow in a polymorphic brackish water threespine stickleback (Gasterosteus aculeatus) population.BMC Evol Biol. 2018 Feb 5;18(1):14. doi: 10.1186/s12862-018-1128-y. BMC Evol Biol. 2018. PMID: 29402230 Free PMC article.
-
Non-parallel divergence across freshwater and marine three-spined stickleback Gasterosteus aculeatus populations.J Fish Biol. 2017 Jul;91(1):175-194. doi: 10.1111/jfb.13336. Epub 2017 May 17. J Fish Biol. 2017. PMID: 28516498
-
Adaptive evolution of lateral plates in three-spined stickleback Gasterosteus aculeatus: a case study in functional analysis of natural variation.J Fish Biol. 2010 Aug;77(2):311-28. doi: 10.1111/j.1095-8649.2010.02640.x. J Fish Biol. 2010. PMID: 20646158 Review.
-
Toward conservation of genetic and phenotypic diversity in Japanese sticklebacks.Genes Genet Syst. 2016 Oct 13;91(2):77-84. doi: 10.1266/ggs.15-00082. Epub 2016 Jun 10. Genes Genet Syst. 2016. PMID: 27301281 Review.
References
-
- Acevedo N, Mercado D, Vergara C, Sánchez J, Kennedy MW, Jiménez S et al. 2009. Association between total immunoglobulin E and antibody responses to naturally acquired Ascaris lumbricoides infection and polymorphisms of immune system-related LIG4, TNFSF13B and IRS2 genes. Clin. Exp. Immunol, 157: 282–290. - PMC - PubMed
-
- Arnold SJ 1992. Constraints on phenotypic evolution. Am. Nat, 140: S85–S107. - PubMed
-
- Barrett RDH, Rogers SM and Schluter D 2008. Natural selection on a major armor gene in threespine stickleback. Science, 322: 255–257. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
