Comprehensive analysis of immune-related biomarkers and pathways in intracerebral hemorrhage using weighted gene co-expression network analysis and competing endogenous ribonucleic acid
- PMID: 36226317
- PMCID: PMC9549172
- DOI: 10.3389/fnmol.2022.955818
Comprehensive analysis of immune-related biomarkers and pathways in intracerebral hemorrhage using weighted gene co-expression network analysis and competing endogenous ribonucleic acid
Abstract
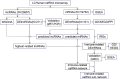
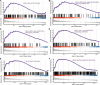
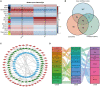
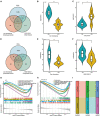
The immune response is an important part of secondary brain injury following intracerebral hemorrhage (ICH), and is related to neurological deficits and prognosis. The mechanisms underlying the immune response and inflammation are of great significance for brain injury and potential functional restoration; however, the immune-related biomarkers and competing endogenous ribonucleic acid (RNA) (ceRNA) networks in the peripheral blood of ICH patients have not yet been constructed. We collected the peripheral blood from ICH patients and controls to assess their ceRNA profiles using LCHuman ceRNA microarray, and to verify their expression with qRT-PCR. Two-hundred-eleven DElncRNAs and one-hundred-one DEmRNAs were detected in the ceRNA microarray of ICH patients. The results of functional enrichment analysis showed that the immune response was an important part of the pathological process of ICH. Twelve lncRNAs, ten miRNAs, and seven mRNAs were present in our constructed immune-related ceRNA network, combining weighted gene co-expression network analysis (WGCNA). Our study was the first to establish the network of the immune-related ceRNAs derived from WGCNA, and to identify leukemia inhibitory factor (LIF) and B cell lymphoma 2-like 13 (BCL2L13) as pivotal immune-related biomarkers in the peripheral blood of ICH patients, which are likely associated with PI3K-Akt, the MAPK signaling pathway, and oxidative phosphorylation. The MOXD2P-miR-211-3p -LIF and LINC00299-miR-198-BCL2L13 axes were indicated to participate in the immune regulatory mechanism of ICH. The goal of our study was to offer innovative insights into the underlying immune regulatory mechanism and to identify possible immune intervention targets for ICH.
Keywords: ICH; WGCNA; ceRNA; immune-related; lncRNA.
Copyright © 2022 Hao, Xu, Wang, Jin, Tang, Zheng, Zhang and He.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Comprehensive Analysis of Aberrantly Expressed Profiles of lncRNAs and miRNAs with Associated ceRNA Network in Lung Adenocarcinoma and Lung Squamous Cell Carcinoma.Pathol Oncol Res. 2020 Jul;26(3):1935-1945. doi: 10.1007/s12253-019-00780-4. Epub 2020 Jan 2. Pathol Oncol Res. 2020. PMID: 31898160
-
Identification of Candidate Blood mRNA Biomarkers in Intracerebral Hemorrhage Using Integrated Microarray and Weighted Gene Co-expression Network Analysis.Front Genet. 2021 Jul 19;12:707713. doi: 10.3389/fgene.2021.707713. eCollection 2021. Front Genet. 2021. PMID: 34349791 Free PMC article.
-
FAM201A, a long noncoding RNA potentially associated with atrial fibrillation identified by ceRNA network analyses and WGCNA.BMC Med Genomics. 2022 Apr 11;15(1):80. doi: 10.1186/s12920-022-01232-w. BMC Med Genomics. 2022. PMID: 35410298 Free PMC article.
-
Reconstruction and Analysis of the Differentially Expressed IncRNA-miRNA-mRNA Network Based on Competitive Endogenous RNA in Hepatocellular Carcinoma.Crit Rev Eukaryot Gene Expr. 2019;29(6):539-549. doi: 10.1615/CritRevEukaryotGeneExpr.2019028740. Crit Rev Eukaryot Gene Expr. 2019. PMID: 32422009 Review.
-
Identification of MFI2-AS1, a Novel Pivotal lncRNA for Prognosis of Stage III/IV Colorectal Cancer.Dig Dis Sci. 2020 Dec;65(12):3538-3550. doi: 10.1007/s10620-020-06064-1. Epub 2020 Jan 20. Dig Dis Sci. 2020. PMID: 31960204 Review.
Cited by
-
Long non-coding RNAs in intracerebral hemorrhage.Front Mol Neurosci. 2023 Jun 12;16:1119275. doi: 10.3389/fnmol.2023.1119275. eCollection 2023. Front Mol Neurosci. 2023. PMID: 37377769 Free PMC article. Review.
-
MiRNAs as potential therapeutic targets and biomarkers for non-traumatic intracerebral hemorrhage.Biomark Res. 2024 Feb 2;12(1):17. doi: 10.1186/s40364-024-00568-y. Biomark Res. 2024. PMID: 38308370 Free PMC article. Review.
-
Epigenetic regulation of the inflammatory response in stroke.Neural Regen Res. 2025 Nov 1;20(11):3045-3062. doi: 10.4103/NRR.NRR-D-24-00672. Epub 2024 Nov 13. Neural Regen Res. 2025. PMID: 39589183 Free PMC article.
References
LinkOut - more resources
Full Text Sources

