Levels of Influenza A Virus Defective Viral Genomes Determine Pathogenesis in the BALB/c Mouse Model
- PMID: 36226985
- PMCID: PMC9645217
- DOI: 10.1128/jvi.01178-22
Levels of Influenza A Virus Defective Viral Genomes Determine Pathogenesis in the BALB/c Mouse Model
Abstract
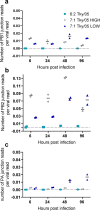
Defective viral genomes (DVGs), which are generated by the viral polymerase in error during RNA replication, can trigger innate immunity and are implicated in altering the clinical outcome of infection. Here, we investigated the impact of DVGs on innate immunity and pathogenicity in a BALB/c mouse model of influenza virus infection. We generated stocks of influenza viruses containing the internal genes of an H5N1 virus that contained different levels of DVGs (indicated by different genome-to-PFU ratios). In lung epithelial cells, the high-DVG stock was immunostimulatory at early time points postinfection. DVGs were amplified during virus replication in myeloid immune cells and triggered proinflammatory cytokine production. In the mouse model, infection with the different virus stocks produced divergent outcomes. The high-DVG stock induced an early type I interferon (IFN) response that limited viral replication in the lungs, resulting in minimal weight loss. In contrast, the virus stock with low levels of DVGs replicated to high titers and amplified DVGs over time, resulting in elevated levels of proinflammatory cytokines accompanied by rapid weight loss and increased morbidity and mortality. Our results suggest that the timing and levels of immunostimulatory DVGs generated during infection contribute to H5N1 pathogenesis. IMPORTANCE Mammalian infections with highly pathogenic avian influenza viruses (HPAIVs) cause severe disease associated with excessive proinflammatory cytokine production. Aberrant replication products, such as defective viral genomes (DVGs), can stimulate the antiviral response, and cytokine induction is associated with their emergence in vivo. We show that stocks of a recombinant virus containing HPAIV internal genes that differ in their amounts of DVGs have vastly diverse outcomes in a mouse model. The high-DVG stock resulted in extremely mild disease due to suppression of viral replication. Conversely, the stock that contained low DVGs but rapidly accumulated DVGs over the course of infection led to severe disease. Therefore, the timing of DVG amplification and proinflammatory cytokine production impact disease outcome, and these findings demonstrate that not all DVG generation reduces viral virulence. This study also emphasizes the crucial requirement to examine the quality of virus preparations regarding DVG content to ensure reproducible research.
Keywords: defective viral genomes; influenza; pathogenesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- de Jong MD, Simmons CP, Thanh TT, Hien VM, Smith GJ, Chau TN, Hoang DM, Chau NV, Khanh TH, Dong VC, Qui PT, Cam BV, Ha DQ, Guan Y, Peiris JS, Chinh NT, Hien TT, Farrar J. 2006. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med 12:1203–1207. 10.1038/nm1477. - DOI - PMC - PubMed
-
- Yu H, Gao Z, Feng Z, Shu Y, Xiang N, Zhou L, Huai Y, Feng L, Peng Z, Li Z, Xu C, Li J, Hu C, Li Q, Xu X, Liu X, Liu Z, Xu L, Chen Y, Luo H, Wei L, Zhang X, Xin J, Guo J, Wang Q, Yuan Z, Zhou L, Zhang K, Zhang W, Yang J, Zhong X, Xia S, Li L, Cheng J, Ma E, He P, Lee SS, Wang Y, Uyeki TM, Yang W. 2008. Clinical characteristics of 26 human cases of highly pathogenic avian influenza A (H5N1) virus infection in China. PLoS One 3:e2985. 10.1371/journal.pone.0002985. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

