Polarly localized WPR proteins interact with PAN receptors and the actin cytoskeleton during maize stomatal development
- PMID: 36227066
- PMCID: PMC9806561
- DOI: 10.1093/plcell/koac301
Polarly localized WPR proteins interact with PAN receptors and the actin cytoskeleton during maize stomatal development
Abstract
Polarization of cells prior to asymmetric cell division is crucial for correct cell divisions, cell fate, and tissue patterning. In maize (Zea mays) stomatal development, the polarization of subsidiary mother cells (SMCs) prior to asymmetric division is controlled by the BRICK (BRK)-PANGLOSS (PAN)-RHO FAMILY GTPASE (ROP) pathway. Two catalytically inactive receptor-like kinases, PAN2 and PAN1, are required for correct division plane positioning. Proteins in the BRK-PAN-ROP pathway are polarized in SMCs, with the polarization of each protein dependent on the previous one. As most of the known proteins in this pathway do not physically interact, possible interactors that might participate in the pathway are yet to be described. We identified WEAK CHLOROPLAST MOVEMENT UNDER BLUE LIGHT 1 (WEB1)/PLASTID MOVEMENT IMPAIRED 2 (PMI2)-RELATED (WPR) proteins as players during SMC polarization in maize. WPRs physically interact with PAN receptors and polarly accumulate in SMCs. The polarized localization of WPR proteins depends on PAN2 but not PAN1. CRISPR-Cas9-induced mutations result in division plane defects in SMCs, and ectopic expression of WPR-RFP results in stomatal defects and alterations to the actin cytoskeleton. We show that certain WPR proteins directly interact with F-actin through their N-terminus. Our data implicate WPR proteins as potentially regulating actin filaments, providing insight into their molecular function. These results demonstrate that WPR proteins are important for cell polarization.
© American Society of Plant Biologists 2022. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures
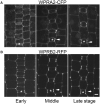



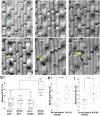
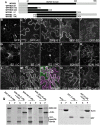

Comment in
-
Forming the queue: A role for WPR proteins in establishing cell polarity.Plant Cell. 2023 Jan 2;35(1):343-344. doi: 10.1093/plcell/koac310. Plant Cell. 2023. PMID: 36282993 Free PMC article. No abstract available.
References
-
- Ashraf MA, Facette M (2020) Plant biology: BASL gives the plant nucleus a sense of direction. Curr Biol 30: R1375–R1377 - PubMed
-
- Campos R, Goff J, Rodriguez-Furlan C, Van Norman JM (2020) The Arabidopsis receptor kinase IRK is polarized and represses specific cell divisions in roots. Dev Cell 52: 183–195 - PubMed
-
- Cartwright HN, Humphries JA, Smith LG (2009) PAN1: a receptor-like protein that promotes polarization of an asymmetric cell division in maize. Science 323: 649–651 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

