Hypermitotic meningiomas harbor DNA methylation subgroups with distinct biological and clinical features
- PMID: 36227281
- PMCID: PMC10013643
- DOI: 10.1093/neuonc/noac224
Hypermitotic meningiomas harbor DNA methylation subgroups with distinct biological and clinical features
Abstract
Background: Meningiomas, the most common primary intracranial tumors, can be separated into 3 DNA methylation groups with distinct biological drivers, clinical outcomes, and therapeutic vulnerabilities. Alternative meningioma grouping schemes using copy number variants, gene expression profiles, somatic short variants, or integrated molecular models have been proposed. These data suggest meningioma DNA methylation groups may harbor subgroups unifying contrasting theories of meningioma biology.
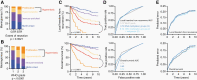
Methods: A total of 565 meningioma DNA methylation profiles from patients with comprehensive clinical follow-up at independent discovery (n = 200) or validation (n = 365) institutions were reanalyzed and classified into Merlin-intact, Immune-enriched, or Hypermitotic DNA methylation groups. RNA sequencing from the discovery (n = 200) or validation (n = 302) cohort were analyzed in the context of DNA methylation groups to identify subgroups. Biological features and clinical outcomes were analyzed across meningioma grouping schemes.
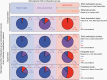
Results: RNA sequencing revealed differential enrichment of FOXM1 target genes across two subgroups of Hypermitotic meningiomas. Differential expression and ontology analyses showed the subgroup of Hypermitotic meningiomas without FOXM1 target gene enrichment was distinguished by gene expression programs driving macromolecular metabolism. Analysis of genetic, epigenetic, gene expression, or cellular features revealed Hypermitotic meningioma subgroups were concordant with Proliferative or Hypermetabolic meningiomas, which were previously reported alongside Merlin-intact and Immune-enriched tumors using an integrated molecular model. The addition of DNA methylation subgroups to clinical models refined the prediction of postoperative outcomes compared to the addition of DNA methylation groups.
Conclusions: Meningiomas can be separated into three DNA methylation groups and Hypermitotic meningiomas can be subdivided into Proliferative and Hypermetabolic subgroups, each with distinct biological and clinical features.
Keywords: DNA methylation; cancer; central nervous system; meningioma; tumor.
© The Author(s) 2022. Published by Oxford University Press on behalf of the Society for Neuro-Oncology. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures


Comment in
-
Bridging deep sequencing to precision oncology in meningiomas.Neuro Oncol. 2023 Mar 14;25(3):531-532. doi: 10.1093/neuonc/noad004. Neuro Oncol. 2023. PMID: 36625527 Free PMC article. No abstract available.
References
-
- Bayley JC, Hadley CC, Harmanci AO, Science AH. Multiple approaches converge on three biological subtypes of meningioma and extract new insights from published studies. Sci Adv. 2022;8(5):eabm6247. https://pubmed.ncbi.nlm.nih.gov/35108039/ - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

