Probing Transient Riboswitch Structures via Single Molecule Accessibility Analysis
- PMID: 36227561
- PMCID: PMC10078578
- DOI: 10.1007/978-1-0716-2687-0_4
Probing Transient Riboswitch Structures via Single Molecule Accessibility Analysis
Abstract
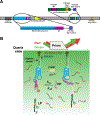
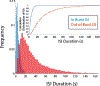
Riboswitches are a class of RNA motifs in the untranslated regions of bacterial messenger RNAs (mRNAs) that can adopt different conformations to regulate gene expression. The binding of specific small molecule or ion ligands, or other RNAs, influences the conformation the riboswitch adopts. Single Molecule Kinetic Analysis of RNA Transient Structure (SiM-KARTS) offers an approach for probing this structural isomerization, or conformational switching, at the level of single mRNA molecules. SiM-KARTS utilizes fluorescently labeled, short, sequence-complementary DNA or RNA oligonucleotide probes that transiently access a specific RNA conformation over another. Binding and dissociation to a surface-immobilized target RNA of arbitrary length are monitored by Total Internal Reflection Fluorescence Microscopy (TIRFM) and quantitatively analyzed, via spike train and burst detection, to elucidate the rate constants of isomerization, revealing mechanistic insights into riboswitching.
Keywords: Bacterial gene regulation; Conformational dynamics; RNA folding; Riboswitch; Single molecule fluorescence microscopy.
© 2023. This is a U.S. government work and not under copyright protection in the U.S.; foreign copyright protection may apply.
Figures
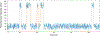
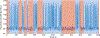
Similar articles
-
Probing RNA structure and interaction dynamics at the single molecule level.Methods. 2019 Jun 1;162-163:3-11. doi: 10.1016/j.ymeth.2019.04.002. Epub 2019 Apr 3. Methods. 2019. PMID: 30951833 Free PMC article. Review.
-
The dynamic nature of RNA as key to understanding riboswitch mechanisms.Acc Chem Res. 2011 Dec 20;44(12):1339-48. doi: 10.1021/ar200035g. Epub 2011 Jun 16. Acc Chem Res. 2011. PMID: 21678902
-
Shine-Dalgarno Accessibility Governs Ribosome Binding to the Adenine Riboswitch.ACS Chem Biol. 2024 Mar 15;19(3):607-618. doi: 10.1021/acschembio.3c00435. Epub 2024 Feb 27. ACS Chem Biol. 2024. PMID: 38412235
-
The Shine-Dalgarno sequence of riboswitch-regulated single mRNAs shows ligand-dependent accessibility bursts.Nat Commun. 2016 Jan 19;7:8976. doi: 10.1038/ncomms9976. Nat Commun. 2016. PMID: 26781350 Free PMC article.
-
Beyond ligand binding: Single molecule observation reveals how riboswitches integrate multiple signals to balance bacterial gene regulation.Curr Opin Struct Biol. 2024 Oct;88:102893. doi: 10.1016/j.sbi.2024.102893. Epub 2024 Jul 26. Curr Opin Struct Biol. 2024. PMID: 39067113 Review.
Cited by
-
Utilizing functional cell-free extracts to dissect ribonucleoprotein complex biology at single-molecule resolution.Wiley Interdiscip Rev RNA. 2023 Sep-Oct;14(5):e1787. doi: 10.1002/wrna.1787. Epub 2023 Apr 12. Wiley Interdiscip Rev RNA. 2023. PMID: 37042458 Free PMC article. Review.
-
A unifying model for microRNA-guided silencing of messenger RNAs.bioRxiv [Preprint]. 2025 Mar 17:2025.03.16.643529. doi: 10.1101/2025.03.16.643529. bioRxiv. 2025. PMID: 40166176 Free PMC article. Preprint.
-
A unifying model for microRNA-guided silencing of messenger RNAs.Res Sq [Preprint]. 2025 Apr 22:rs.3.rs-6422368. doi: 10.21203/rs.3.rs-6422368/v1. Res Sq. 2025. PMID: 40313740 Free PMC article. Preprint.
References
-
- Akimoto H, Imamiya E, Hitaka T, Nomura H, & Nishimura S (1988). Synthesis of queuine, the base of naturally occurring hypermodified nucleoside (queuosine), and its analogues. Journal of the Chemical Society, Perkin Transactions 1(7), 1637–1644. doi:10.1039/P19880001637 - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

