The wtf meiotic driver gene family has unexpectedly persisted for over 100 million years
- PMID: 36227631
- PMCID: PMC9562144
- DOI: 10.7554/eLife.81149
The wtf meiotic driver gene family has unexpectedly persisted for over 100 million years
Abstract
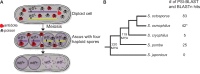
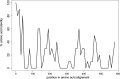
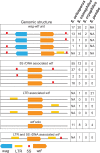
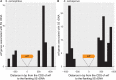
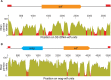
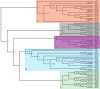
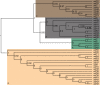
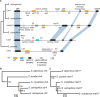
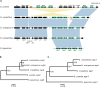
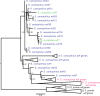
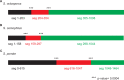
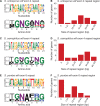
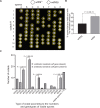
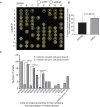
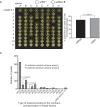
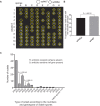
Meiotic drivers are selfish elements that bias their own transmission into more than half of the viable progeny produced by a driver+/driver- heterozygote. Meiotic drivers are thought to exist for relatively short evolutionary timespans because a driver gene or gene family is often found in a single species or in a group of very closely related species. Additionally, drivers are generally considered doomed to extinction when they spread to fixation or when suppressors arise. In this study, we examine the evolutionary history of the wtf meiotic drivers first discovered in the fission yeast Schizosaccharomyces pombe. We identify homologous genes in three other fission yeast species, S. octosporus, S. osmophilus, and S. cryophilus, which are estimated to have diverged over 100 million years ago from the S. pombe lineage. Synteny evidence supports that wtf genes were present in the common ancestor of these four species. Moreover, the ancestral genes were likely drivers as wtf genes in S. octosporus cause meiotic drive. Our findings indicate that meiotic drive systems can be maintained for long evolutionary timespans.
Keywords: S. octosporus; S. pombe; evolutionary biology; genetics; genomics; meiosis; meiotic drive; selfish genes; spore killers; wtf.
© 2022, De Carvalho, Jia et al.
Conflict of interest statement
MD, GJ, AN, RB, YX, JL, IS, LD, SZ No competing interests declared
Figures











References
-
- Akaike H. Information Theory and an Extension of the Maximum Likelihood Principle. Springer; 1998. - DOI
Publication types
MeSH terms
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

