The Role of the Environment in Horizontal Gene Transfer
- PMID: 36227733
- PMCID: PMC9641970
- DOI: 10.1093/molbev/msac220
The Role of the Environment in Horizontal Gene Transfer
Abstract
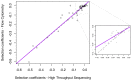
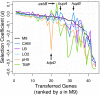
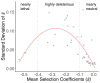
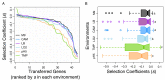
Gene-by-environment interactions play a crucial role in horizontal gene transfer by affecting how the transferred genes alter host fitness. However, how the environment modulates the fitness effect of transferred genes has not been tested systematically in an experimental study. We adapted a high-throughput technique for obtaining very precise estimates of bacterial fitness, in order to measure the fitness effects of 44 orthologs transferred from Salmonella Typhimurium to Escherichia coli in six physiologically relevant environments. We found that the fitness effects of individual genes were highly dependent on the environment, while the distributions of fitness effects across genes were not, with all tested environments resulting in distributions of same shape and spread. Furthermore, the extent to which the fitness effects of a gene varied between environments depended on the average fitness effect of that gene across all environments, with nearly neutral and nearly lethal genes having more consistent fitness effects across all environments compared to deleterious genes. Put together, our results reveal the unpredictable nature of how environmental conditions impact the fitness effects of each individual gene. At the same time, distributions of fitness effects across environments exhibit consistent features, pointing to the generalizability of factors that shape horizontal gene transfer of orthologous genes.
Keywords: distribution of fitness effects; evolutionary barriers; gene-by-environment interactions; horizontal gene transfer.
© The Author(s) 2022. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures
References
-
- Amaral L, Fanning S, Pagès JM. 2011. Efflux pumps of gram-negative bacteria: genetic responses to stress and the modulation of their activity by pH, inhibitors, and phenothiazines. Adv Enzymol Relat Areas Mol Biol. 77:61–108. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

