Molecular convergence by differential domain acquisition is a hallmark of chromosomal passenger complex evolution
- PMID: 36227914
- PMCID: PMC9680938
- DOI: 10.1073/pnas.2200108119
Molecular convergence by differential domain acquisition is a hallmark of chromosomal passenger complex evolution
Abstract
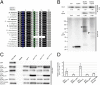
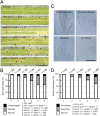
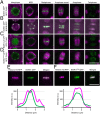
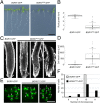
The chromosomal passenger complex (CPC) is a heterotetrameric regulator of eukaryotic cell division, consisting of an Aurora-type kinase and a scaffold built of INCENP, Borealin, and Survivin. While most CPC components are conserved across eukaryotes, orthologs of the chromatin reader Survivin have previously only been found in animals and fungi, raising the question of how its essential role is carried out in other eukaryotes. By characterizing proteins that bind to the Arabidopsis Borealin ortholog, we identified BOREALIN RELATED INTERACTOR 1 and 2 (BORI1 and BORI2) as redundant Survivin-like proteins in the context of the CPC in plants. Loss of BORI function is lethal and a reduced expression of BORIs causes severe developmental defects. Similar to Survivin, we find that the BORIs bind to phosphorylated histone H3, relevant for correct CPC association with chromatin. However, this interaction is not mediated by a BIR domain as in previously recognized Survivin orthologs but by an FHA domain, a widely conserved phosphate-binding module. We find that the unifying criterion of Survivin-type proteins is a helix that facilitates complex formation with the other two scaffold components and that the addition of a phosphate-binding domain, necessary for concentration at the inner centromere, evolved in parallel in different eukaryotic groups. Using sensitive similarity searches, we find conservation of this helical domain between animals and plants and identify the missing CPC component in most eukaryotic supergroups. Interestingly, we also detect Survivin orthologs without a defined phosphate-binding domain, likely reflecting the situation in the last eukaryotic common ancestor.
Keywords: cell division; evolution; microtuble cytoskeleton.
Conflict of interest statement
The authors declare no competing interest.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

