Insight into the regulatory networks underlying the high lipid perennial ryegrass growth under different irradiances
- PMID: 36227922
- PMCID: PMC9560171
- DOI: 10.1371/journal.pone.0275503
Insight into the regulatory networks underlying the high lipid perennial ryegrass growth under different irradiances
Abstract
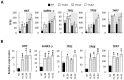
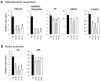
Under favourable conditions, perennial ryegrass (Lolium perenne) engineered to accumulated high lipid (HL) carbon sink in their leaves was previously shown to also enhance photosynthesis and growth. The greater aboveground biomass was found to be diminished in a dense canopy compared to spaced pots. Besides, the underlying genetic regulatory network linking between leaf lipid sinks and these physiological changes remains unknown. In this study, we demonstrated that the growth advantage was not displayed in HL Lolium grown in spaced pots under low lights. Under standard lights, analysis of differentiating transcripts in HL Lolium reveals that the plants had elevated transcripts involved in lipid metabolism, light capturing, photosynthesis, and sugar signalling while reduced expression of genes participating in sugar biosynthesis and transportation. The plants also had altered several transcripts involved in mitochondrial oxidative respiration and redox potential. Many of the above upregulated or downregulated transcript levels were found to be complemented by growing the plants under low light. Overall, this study emphasizes the importance of carbon and energy homeostatic regulatory mechanisms to overall productivity of the HL Lolium through photosynthesis, most of which are significantly impacted by low irradiances.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

