Discovery of Schistosoma mekongi circulating proteins and antigens in infected mouse sera
- PMID: 36227939
- PMCID: PMC9562170
- DOI: 10.1371/journal.pone.0275992
Discovery of Schistosoma mekongi circulating proteins and antigens in infected mouse sera
Abstract
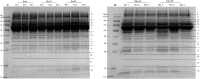
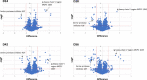
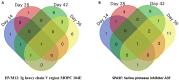
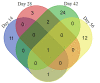
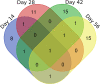
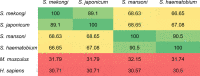
Schistosomiasis is a neglected tropical disease caused by an infection of the parasitic flatworms schistosomes. Schistosoma mekongi is a restricted Schistosoma species found near the Mekong River, mainly in southern Laos and northern Cambodia. Because there is no vaccine or effective early diagnosis available for S. mekongi, additional biomarkers are required. In this study, serum biomarkers associated with S. mekongi-infected mice were identified at 14-, 28-, 42-, and 56-days post-infection. Circulating proteins and antigens of S. mekongi in mouse sera were analyzed using mass spectrometry-based proteomics. Serine protease inhibitors and macrophage erythroblast attacher were down-regulated in mouse sera at all infection timepoints. In addition, 54 circulating proteins and 55 antigens of S. mekongi were identified. Notable circulating proteins included kyphoscoliosis peptidase and putative tuberin, and antigens were detected at all four infection timepoints, particularly in the early stages (12 days). The putative tuberin sequence of S. mekongi was highly similar to homologs found in other members of the genus Schistosoma and less similar to human and murine sequences. Our study provided the identity of promising diagnostic biomarkers that could be applicable in early schistosomiasis diagnosis and vaccine development.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Schistosoma mekongi cathepsin B and its use in the development of an immunodiagnosis.Acta Trop. 2016 Mar;155:11-9. doi: 10.1016/j.actatropica.2015.11.017. Epub 2015 Dec 2. Acta Trop. 2016. PMID: 26655041
-
Proteomic and immunomic analysis of Schistosoma mekongi egg proteins.Exp Parasitol. 2018 Aug;191:88-96. doi: 10.1016/j.exppara.2018.07.002. Epub 2018 Jul 17. Exp Parasitol. 2018. PMID: 30009810
-
Identification of Low Molecular Weight Proteins and Peptides from Schistosoma mekongi Worm, Egg and Infected Mouse Sera.Biomolecules. 2021 Apr 11;11(4):559. doi: 10.3390/biom11040559. Biomolecules. 2021. PMID: 33920436 Free PMC article.
-
Schistosomiasis mekongi: from discovery to control.Parasitol Int. 2004 Jun;53(2):135-42. doi: 10.1016/j.parint.2004.01.004. Parasitol Int. 2004. PMID: 15081945 Review.
-
Current advances in serological and molecular diagnosis of Schistosoma mekongi infection.Trop Med Health. 2024 Apr 22;52(1):32. doi: 10.1186/s41182-024-00598-0. Trop Med Health. 2024. PMID: 38650044 Free PMC article. Review.
Cited by
-
Chromosome-scale genome of the human blood fluke Schistosoma mekongi and its implications for public health.Infect Dis Poverty. 2023 Nov 28;12(1):104. doi: 10.1186/s40249-023-01160-6. Infect Dis Poverty. 2023. PMID: 38017557 Free PMC article.
-
A Place-Based Conceptual Model (PBCM) of Neotricula aperta/Schistosoma mekongi habitat before and after dam construction in the Lower Mekong River.PLoS Negl Trop Dis. 2023 Oct 6;17(10):e0011122. doi: 10.1371/journal.pntd.0011122. eCollection 2023 Oct. PLoS Negl Trop Dis. 2023. PMID: 37801463 Free PMC article.
-
Micro-scale genetic structure and genetic variation of Neotricula aperta (Gastropoda: Pomatiopsidae), the intermediate host of Schistosoma mekongi (Digenea: Schistosomatidae) in Champasak Province, Laos.Trop Med Health. 2025 Jul 24;53(1):97. doi: 10.1186/s41182-025-00775-9. Trop Med Health. 2025. PMID: 40708054 Free PMC article.
-
Global distribution of zoonotic digenetic trematodes: a scoping review.Infect Dis Poverty. 2024 Jun 14;13(1):46. doi: 10.1186/s40249-024-01208-1. Infect Dis Poverty. 2024. PMID: 38877531 Free PMC article.
-
Identification of biomarker candidates for filarial parasite infections by analysis of extracellular vesicles.Front Parasitol. 2023 Oct 23;2:1281092. doi: 10.3389/fpara.2023.1281092. eCollection 2023. Front Parasitol. 2023. PMID: 39816829 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

