The choice of the objective function in flux balance analysis is crucial for predicting replicative lifespans in yeast
- PMID: 36227951
- PMCID: PMC9560524
- DOI: 10.1371/journal.pone.0276112
The choice of the objective function in flux balance analysis is crucial for predicting replicative lifespans in yeast
Abstract
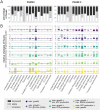
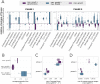
Flux balance analysis (FBA) is a powerful tool to study genome-scale models of the cellular metabolism, based on finding the optimal flux distributions over the network. While the objective function is crucial for the outcome, its choice, even though motivated by evolutionary arguments, has not been directly connected to related measures. Here, we used an available multi-scale mathematical model of yeast replicative ageing, integrating cellular metabolism, nutrient sensing and damage accumulation, to systematically test the effect of commonly used objective functions on features of replicative ageing in budding yeast, such as the number of cell divisions and the corresponding time between divisions. The simulations confirmed that assuming maximal growth is essential for reaching realistic lifespans. The usage of the parsimonious solution or the additional maximisation of a growth-independent energy cost can improve lifespan predictions, explained by either increased respiratory activity using resources otherwise allocated to cellular growth or by enhancing antioxidative activity, specifically in early life. Our work provides a new perspective on choosing the objective function in FBA by connecting it to replicative ageing.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Estimating network changes from lifespan measurements using a parsimonious gene network model of cellular aging.BMC Bioinformatics. 2019 Nov 20;20(1):599. doi: 10.1186/s12859-019-3177-7. BMC Bioinformatics. 2019. PMID: 31747877 Free PMC article.
-
Multi-scale model suggests the trade-off between protein and ATP demand as a driver of metabolic changes during yeast replicative ageing.PLoS Comput Biol. 2022 Jul 7;18(7):e1010261. doi: 10.1371/journal.pcbi.1010261. eCollection 2022 Jul. PLoS Comput Biol. 2022. PMID: 35797415 Free PMC article.
-
Preadaptation to efficient respiratory maintenance is essential both for maximal longevity and the retention of replicative potential in chronologically ageing yeast.Mech Ageing Dev. 2006 Sep;127(9):733-40. doi: 10.1016/j.mad.2006.05.004. Epub 2006 Jun 19. Mech Ageing Dev. 2006. PMID: 16784770
-
Long-lived yeast as a model for ageing research.Yeast. 2006 Feb;23(3):215-26. doi: 10.1002/yea.1354. Yeast. 2006. PMID: 16498698 Review.
-
Longevity pathways and maintenance of the proteome: the role of autophagy and mitophagy during yeast ageing.Microb Cell. 2014 Apr 7;1(4):118-127. doi: 10.15698/mic2014.04.136. Microb Cell. 2014. PMID: 28357232 Free PMC article. Review.
Cited by
-
Flux sampling in genome-scale metabolic modeling of microbial communities.BMC Bioinformatics. 2024 Jan 29;25(1):45. doi: 10.1186/s12859-024-05655-3. BMC Bioinformatics. 2024. PMID: 38287239 Free PMC article.
-
Model validation and selection in metabolic flux analysis and flux balance analysis.Biotechnol Prog. 2024 Jan-Feb;40(1):e3413. doi: 10.1002/btpr.3413. Epub 2023 Nov 24. Biotechnol Prog. 2024. PMID: 37997613 Free PMC article. Review.
-
Flux Sampling in Genome-scale Metabolic Modeling of Microbial Communities.bioRxiv [Preprint]. 2023 Apr 20:2023.04.18.537368. doi: 10.1101/2023.04.18.537368. bioRxiv. 2023. Update in: BMC Bioinformatics. 2024 Jan 29;25(1):45. doi: 10.1186/s12859-024-05655-3. PMID: 37197028 Free PMC article. Updated. Preprint.
-
Metabolic Objectives and Trade-Offs: Inference and Applications.Metabolites. 2025 Feb 6;15(2):101. doi: 10.3390/metabo15020101. Metabolites. 2025. PMID: 39997726 Free PMC article. Review.
-
Discretised Flux Balance Analysis for Reaction-Diffusion Simulation of Single-Cell Metabolism.Bull Math Biol. 2024 Mar 6;86(4):39. doi: 10.1007/s11538-024-01264-6. Bull Math Biol. 2024. PMID: 38448618 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

