Sentinel p16INK4a+ cells in the basement membrane form a reparative niche in the lung
- PMID: 36227993
- PMCID: PMC10621323
- DOI: 10.1126/science.abf3326
Sentinel p16INK4a+ cells in the basement membrane form a reparative niche in the lung
Abstract
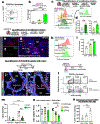
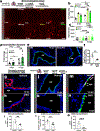
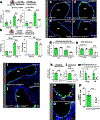
We engineered an ultrasensitive reporter of p16INK4a, a biomarker of cellular senescence. Our reporter detected p16INK4a-expressing fibroblasts with certain senescent characteristics that appeared shortly after birth in the basement membrane adjacent to epithelial stem cells in the lung. Furthermore, these p16INK4a+ fibroblasts had enhanced capacity to sense tissue inflammation and respond through their increased secretory capacity to promote epithelial regeneration. In addition, p16INK4a expression was required in fibroblasts to enhance epithelial regeneration. This study highlights a role for p16INK4a+ fibroblasts as tissue-resident sentinels in the stem cell niche that monitor barrier integrity and rapidly respond to inflammation to promote tissue regeneration.
Conflict of interest statement
Figures



Comment in
-
Senescent fibroblasts support lung repair.Nat Rev Mol Cell Biol. 2022 Dec;23(12):775. doi: 10.1038/s41580-022-00559-7. Nat Rev Mol Cell Biol. 2022. PMID: 36329214 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

