Data-independent acquisition and quantification of extracellular matrix from human lung in chronic inflammation-associated carcinomas
- PMID: 36228107
- PMCID: PMC10391693
- DOI: 10.1002/pmic.202200021
Data-independent acquisition and quantification of extracellular matrix from human lung in chronic inflammation-associated carcinomas
Abstract
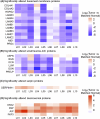
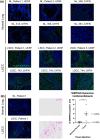
Early events associated with chronic inflammation and cancer involve significant remodeling of the extracellular matrix (ECM), which greatly affects its composition and functional properties. Using lung squamous cell carcinoma (LSCC), a chronic inflammation-associated cancer (CIAC), we optimized a robust proteomic pipeline to discover potential biomarker signatures and protein changes specifically in the stroma. We combined ECM enrichment from fresh human tissues, data-independent acquisition (DIA) strategies, and stringent statistical processing to analyze "Tumor" and matched adjacent histologically normal ("Matched Normal") tissues from patients with LSCC. Overall, 1802 protein groups were quantified with at least two unique peptides, and 56% of those proteins were annotated as "extracellular." Confirming dramatic ECM remodeling during CIAC progression, 529 proteins were significantly altered in the "Tumor" compared to "Matched Normal" tissues. The signature was typified by a coordinated loss of basement membrane proteins and small leucine-rich proteins. The dramatic increase in the stromal levels of SERPINH1/heat shock protein 47, that was discovered using our ECM proteomic pipeline, was validated by immunohistochemistry (IHC) of "Tumor" and "Matched Normal" tissues, obtained from an independent cohort of LSCC patients. This integrated workflow provided novel insights into ECM remodeling during CIAC progression, and identified potential biomarker signatures and future therapeutic targets.
Keywords: data-independent acquisition; extracellular matrix; lung squamous cell carcinoma; quantification; serpins.
© 2022 The Authors. Proteomics published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Hecht, S. S. (2011). Tobacco smoke carcinogens and lung cancer. In Penning T. M. (Ed.), Chemical carcinogenesis (pp. 53–74). Humana Press.
-
- Tsao, A. S. , Liu, D. , Lee, J. J. , Spitz, M. , & Hong, W. K. (2006). Smoking affects treatment outcome in patients with advanced nonsmall cell lung cancer. Cancer, 106(11), 2428–2436. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

