Human leukocyte antigen alleles associate with COVID-19 vaccine immunogenicity and risk of breakthrough infection
- PMID: 36228659
- PMCID: PMC9873562
- DOI: 10.1038/s41591-022-02078-6
Human leukocyte antigen alleles associate with COVID-19 vaccine immunogenicity and risk of breakthrough infection
Abstract
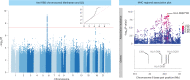
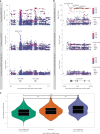
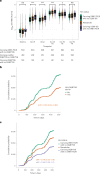
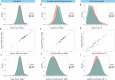
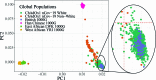
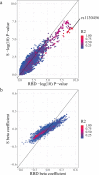
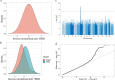
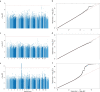
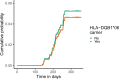
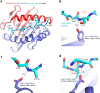
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) vaccine immunogenicity varies between individuals, and immune responses correlate with vaccine efficacy. Using data from 1,076 participants enrolled in ChAdOx1 nCov-19 vaccine efficacy trials in the United Kingdom, we found that inter-individual variation in normalized antibody responses against SARS-CoV-2 spike and its receptor-binding domain (RBD) at 28 days after first vaccination shows genome-wide significant association with major histocompatibility complex (MHC) class II alleles. The most statistically significant association with higher levels of anti-RBD antibody was HLA-DQB1*06 (P = 3.2 × 10-9), which we replicated in 1,677 additional vaccinees. Individuals carrying HLA-DQB1*06 alleles were less likely to experience PCR-confirmed breakthrough infection during the ancestral SARS-CoV-2 virus and subsequent Alpha variant waves compared to non-carriers (hazard ratio = 0.63, 0.42-0.93, P = 0.02). We identified a distinct spike-derived peptide that is predicted to bind differentially to HLA-DQB1*06 compared to other similar alleles, and we found evidence of increased spike-specific memory B cell responses in HLA-DQB1*06 carriers at 84 days after first vaccination. Our results demonstrate association of HLA type with Coronavirus Disease 2019 (COVID-19) vaccine antibody response and risk of breakthrough infection, with implications for future vaccine design and implementation.
© 2023. The Author(s).
Conflict of interest statement
The University of Oxford has entered into a partnership with AstraZeneca for further development of ChAdOx1 nCoV-19. T.L. is named as an inventor on a patent application covering this SARS-CoV-2 vaccine and was a consultant to Vaccitech for an unrelated project. A.J.P. is Chair of the UK Department of Health and Social Care (DHSC) Joint Committee on Vaccination & Immunisation (JCVI) but does not participate in discussions on COVID-19 vaccines and is a member of the Strategic Advisory Group of Experts on Immunization to the World Health Organization (WHO). A.J.P. is an NIHR senior investigator. The remaining authors declare no competing interests. The views expressed in this article do not necessarily represent the views of the DHSC, JCVI, NIHR or WHO.
Figures



References
-
- COVID-19 Map. Johns Hopkins Coronavirus Resource Center. https://coronavirus.jhu.edu/map.html
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

