Quindoline-derivatives display potent G-quadruplex-mediated antiviral activity against herpes simplex virus 1
- PMID: 36228762
- PMCID: PMC9720158
- DOI: 10.1016/j.antiviral.2022.105432
Quindoline-derivatives display potent G-quadruplex-mediated antiviral activity against herpes simplex virus 1
Abstract
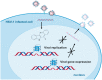
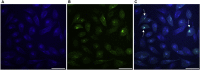
G-quadruplexes (G4s) are non-canonical nucleic acid structures that regulate key biological processes, from transcription to genome replication both in humans and viruses. The herpes simplex virus-1 (HSV-1) genome is prone to form G4s that, along with proteins, regulate its viral cycle. General G4 ligands have been shown to hamper the viral cycle, pointing to viral G4s as original antiviral targets. Because cellular G4s are also normally present in infected cells, the quest for improved anti-HSV-1 G4 ligands is still open. Here, we evaluated a series of new quindoline-derivatives which showed high binding to and stabilization of the viral G4s. They displayed nanomolar-range anti-HSV-1 activity paralleled by negligible cytotoxicity in human cells, thus proving remarkable selectivity. The best-in-class compound inhibited the viral life cycle at the early times post infection up to the step of viral genome replication. In infected human cells, it reduced expression of ICP4, the main viral transcription factor, by stabilizing the G4s embedded in ICP4 promoter. Quindoline-derivatives thus emerge as a new class of G4 ligands with potent dual anti HSV-1 activity.
Keywords: Antiviral activity; G-quadruplex; HSV-1; ICP4; Quindoline derivatives.
Copyright © 2022 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




References
-
- Alfonso-Dunn R., Turner A.-M.W., Beltran P.M.J., Arbuckle J.H., Budayeva H.G., Cristea I.M., Kristie T.M. Transcriptional elongation of HSV Immediate Early genes by the Super Elongation Complex drives lytic infection and reactivation from latency. Cell Host Microbe. 2017;21:507–517. doi: 10.1016/j.chom.2017.03.007. e5. - DOI - PMC - PubMed
-
- Artusi S., Nadai M., Perrone R., Biasolo M.A., Palù G., Flamand L., Calistri A., Richter S.N. The Herpes Simplex Virus-1 genome contains multiple clusters of repeated G-quadruplex: implications for the antiviral activity of a G-quadruplex ligand. Antivir. Res. 2015;118:123–131. doi: 10.1016/j.antiviral.2015.03.016. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

