Whole-exome sequencing of epithelial ovarian carcinomas differing in resistance to platinum therapy
- PMID: 36229065
- PMCID: PMC9574568
- DOI: 10.26508/lsa.202201551
Whole-exome sequencing of epithelial ovarian carcinomas differing in resistance to platinum therapy
Abstract
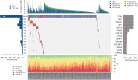
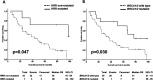
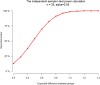
Epithelial ovarian carcinoma (EOC) is highly fatal because of the risk of resistance to therapy and recurrence. We performed whole-exome sequencing of blood and tumor tissue pairs of 50 patients with surgically resected EOC. Compared with sensitive patients, platinum-resistant patients had a significantly higher somatic mutational rate in <i>TP53</i> and lower in several genes from the Hippo pathway. We confirmed the pivotal role of somatic mutations in homologous recombination repair genes in platinum sensitivity and favorable prognosis of EOC patients. Implementing the germline homologous recombination repair profile significantly improved the prediction. In addition, distinct mutational signatures, for example, SBS6, and overall mutational load, somatic mutations in <i>PABPC1</i>, <i>PABPC3</i>, and <i>TFAM</i> co-segregated with the resistance status, high-grade serous carcinoma subtype, or overall survival of patients. We generated germline and somatic genetic landscapes of prognostically different subgroups of EOC patients for further follow-up studies focused on utilizing the observed associations in precision oncology.
© 2022 Hlaváč et al.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures















References
-
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc B Methodol 57: 289–300. 10.1111/j.2517-6161.1995.tb02031.x - DOI
-
- Brachova P, Mueting SR, Carlson MJ, Goodheart MJ, Button AM, Mott SL, Dai D, Thiel KW, Devor EJ, Leslie KK (2015) TP53 oncomorphic mutations predict resistance to platinum- and taxane-based standard chemotherapy in patients diagnosed with advanced serous ovarian carcinoma. Int J Oncol 46: 607–618. 10.3892/ijo.2014.2747 - DOI - PMC - PubMed
-
- Broad Institute TCGA Genome Data Analysis Center (2016) Mutation Analysis (MutSig 2CV v3.1). Broad Institute of MIT and Harvard. 10.7908/C1736QC5 Accessed May 25, 2022. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
