A dominant negative mitofusin causes mitochondrial perinuclear clusters because of aberrant tethering
- PMID: 36229071
- PMCID: PMC9568670
- DOI: 10.26508/lsa.202101305
A dominant negative mitofusin causes mitochondrial perinuclear clusters because of aberrant tethering
Abstract
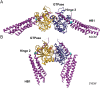
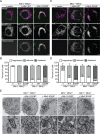
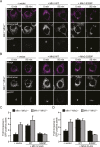
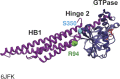
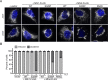
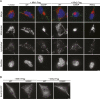
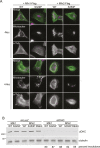
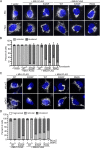
In vertebrates, mitochondrial outer membrane fusion is mediated by two mitofusin paralogs, Mfn1 and Mfn2, conserved dynamin superfamily proteins. Here, we characterize a variant of mitofusin reported in patients with CMT2A where a serine is replaced with a proline (Mfn2-S350P and the equivalent in Mfn1, S329P). This serine is in a hinge domain (Hinge 2) that connects the globular GTPase domain to the adjacent extended helical bundle. We find that expression of this variant results in prolific and stable mitochondrial tethering that also blocks mitochondrial fusion by endogenous wild-type mitofusin. The formation of mitochondrial perinuclear clusters by this CMT2A variant requires normal GTPase domain function and formation of a mitofusin complex across two membranes. We propose that conformational dynamics mediated by Hinge 2 and regulated by GTP hydrolysis are disrupted by the substitution of proline at S329/S350 and this prevents progression from tethering to membrane fusion. Thus, our data are consistent with a model for mitofusin-mediated membrane fusion where Hinge 2 supports a power stroke to progress from the tethering complex to membrane fusion.
© 2022 Sloat and Hoppins.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures






References
-
- Anton V, Buntenbroich I, Schuster R, Babatz F, Simões T, Altin S, Calabrese G, Riemer J, Schauss A, Escobar-Henriques M (2019) Plasticity in salt bridge allows fusion-competent ubiquitylation of mitofusins and Cdc48 recognition. Life Sci Alliance 2: e201900491. 10.26508/lsa.201900491 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
