Brain de novo transcriptome assembly of a toad species showing polymorphic anti-predatory behavior
- PMID: 36229462
- PMCID: PMC9561626
- DOI: 10.1038/s41597-022-01724-5
Brain de novo transcriptome assembly of a toad species showing polymorphic anti-predatory behavior
Abstract
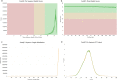
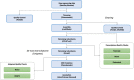
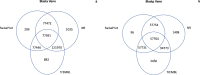
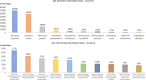
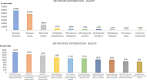
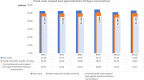
Understanding the genomic underpinnings of antipredatory behaviors is a hot topic in eco-evolutionary research. Yellow-bellied toad of the genus Bombina are textbook examples of the deimatic display, a time-structured behavior aimed at startling predators. Here, we generated the first de novo brain transcriptome of the Apennine yellow-bellied toad Bombina pachypus, a species showing inter-individual variation in the deimatic display. Through Rna-Seq experiments on a set of individuals showing distinct behavioral phenotypes, we generated 316,329,573 reads, which were assembled and annotated. The high-quality assembly was confirmed by assembly validators and by aligning the contigs against the de novo transcriptome with a mapping percentage higher than 91.0%. The homology annotation with DIAMOND (blastx) led to 77,391 contigs annotated on Nr, Swiss Prot and TrEMBL, whereas the domain and site protein prediction made with InterProScan led to 4747 GO-annotated and 1025 KEGG-annotated contigs. The B. pachypus transcriptome described here will be a valuable resource for further studies on the genomic underpinnings of behavioral variation in amphibians.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
De novo transcriptome assembly and annotation for gene discovery in Salamandra salamandra at the larval stage.Sci Data. 2023 May 27;10(1):330. doi: 10.1038/s41597-023-02217-9. Sci Data. 2023. PMID: 37244908 Free PMC article.
-
RNA-Seq analysis and transcriptome assembly for blackberry (Rubus sp. Var. Lochness) fruit.BMC Genomics. 2015 Jan 22;16(1):5. doi: 10.1186/s12864-014-1198-1. BMC Genomics. 2015. PMID: 25608670 Free PMC article.
-
De novo assembly and annotation of the marine mysid (Neomysis awatschensis) transcriptome.Mar Genomics. 2016 Aug;28:41-43. doi: 10.1016/j.margen.2016.05.001. Epub 2016 May 14. Mar Genomics. 2016. PMID: 27189440
-
A Comparison of Resources for the Annotation of a De Novo Assembled Transcriptome in the Molting Gland (Y-Organ) of the Blackback Land Crab, Gecarcinus lateralis.Integr Comp Biol. 2016 Dec;56(6):1103-1112. doi: 10.1093/icb/icw107. Epub 2016 Aug 22. Integr Comp Biol. 2016. PMID: 27549198
-
A simple guide to de novo transcriptome assembly and annotation.Brief Bioinform. 2022 Mar 10;23(2):bbab563. doi: 10.1093/bib/bbab563. Brief Bioinform. 2022. PMID: 35076693 Free PMC article. Review.
Cited by
-
HPC-T-Assembly: a pipeline for de novo transcriptome assembly of large multi-specie datasets.BMC Bioinformatics. 2025 Apr 28;26(1):113. doi: 10.1186/s12859-025-06121-4. BMC Bioinformatics. 2025. PMID: 40295976 Free PMC article.
-
HPC-T-Annotator: an HPC tool for de novo transcriptome assembly annotation.BMC Bioinformatics. 2024 Aug 21;25(1):272. doi: 10.1186/s12859-024-05887-3. BMC Bioinformatics. 2024. PMID: 39169276 Free PMC article.
-
De novo transcriptome assembly of an Antarctic nematode for the study of thermal adaptation in marine parasites.Sci Data. 2023 Oct 19;10(1):720. doi: 10.1038/s41597-023-02591-4. Sci Data. 2023. PMID: 37857654 Free PMC article.
-
De novo transcriptome assembly of the Mediterranean sea-rock pool mosquitoes Aedes mariae and Aedes zammitii.Sci Data. 2025 Jan 20;12(1):115. doi: 10.1038/s41597-025-04393-2. Sci Data. 2025. PMID: 39833234 Free PMC article.
-
De novo transcriptome assembly and annotation for gene discovery in Salamandra salamandra at the larval stage.Sci Data. 2023 May 27;10(1):330. doi: 10.1038/s41597-023-02217-9. Sci Data. 2023. PMID: 37244908 Free PMC article.
References
-
- Carere, C. & Maestripieri, D. Animal Personalities: Behavior, Physiology, and Evolution. (Chicago: University of Chicago Press, 2013).
-
- Jensen P. Behaviour epigenetics–the connection between environment, stress and welfare. Appl. Anim. Behav. Sci. 2014;157:1–7. doi: 10.1016/j.applanim.2014.02.009. - DOI
-
- Van Oers K, de Jong G, van Noordwijk AJ, Kempenaers B, Drent PJ. Contribution of genetics to the study of animal personalities: a review of case studies. Behaviour. 2005;142:1185–120610. doi: 10.1163/156853905774539364. - DOI
-
- Van Oers, K. & Sinn, D. L. The quantitative and molecular genetics of animal personality. In: Carere, C. & Maestripieri, D. editors. Animal Personalities: Behavior, Physiology, and Evolution. (Chicago: University of Chicago Press; p. 148–200, 2013).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

