Spatiotemporal variations in retrovirus-host interactions among Darwin's finches
- PMID: 36229469
- PMCID: PMC9562234
- DOI: 10.1038/s41467-022-33723-w
Spatiotemporal variations in retrovirus-host interactions among Darwin's finches
Abstract
Endogenous retroviruses (ERVs) are inherited remnants of retroviruses that colonized host germline over millions of years, providing a sampling of retroviral diversity across time. Here, we utilize the strength of Darwin's finches, a system synonymous with evolutionary studies, for investigating ERV history, revealing recent retrovirus-host interactions in natural populations. By mapping ERV variation across all species of Darwin's finches and comparing with outgroup species, we highlight geographical and historical patterns of retrovirus-host occurrence, utilizing the system for evaluating the extent and timing of retroviral activity in hosts undergoing adaptive radiation and colonization of new environments. We find shared ERVs among all samples indicating retrovirus-host associations pre-dating host speciation, as well as considerable ERV variation across populations of the entire Darwin's finches' radiation. Unexpected ERV variation in finch species on different islands suggests historical changes in gene flow and selection. Non-random distribution of ERVs along and between chromosomes, and across finch species, suggests association between ERV accumulation and the rapid speciation of Darwin's finches.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

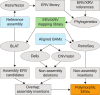

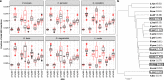
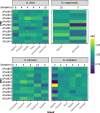
References
-
- Grant, P. R. Ecology and evolution of Darwin’s finches. (Princeton University Press, 1986).
-
- Funk ER, Burns KJ. Biogeographic origins of Darwin’s finches (Thraupidae: Coerebinae) Auk. 2018;135:561–571. doi: 10.1642/AUK-17-215.1. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

