Assessment of variability in the plasma 7k SomaScan proteomics assay
- PMID: 36229504
- PMCID: PMC9561184
- DOI: 10.1038/s41598-022-22116-0
Assessment of variability in the plasma 7k SomaScan proteomics assay
Abstract
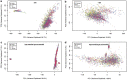
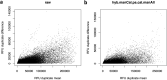
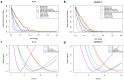
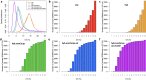
SomaScan is a high-throughput, aptamer-based proteomics assay designed for the simultaneous measurement of thousands of proteins with a broad range of endogenous concentrations. In its most current version, the 7k SomaScan assay v4.1 is capable of measuring 7288 human proteins. In this work, we present an extensive technical assessment of this platform based on a study of 2050 samples across 22 plates. Included in the study design were inter-plate technical duplicates from 102 human subjects, which allowed us to characterize different normalization procedures, evaluate assay variability by multiple analytical approaches, present signal-over-background metrics, and discuss potential specificity issues. By providing detailed performance assessments on this wide range of technical aspects, we aim for this work to serve as a valuable resource for the growing community of SomaScan users.
© 2022. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Schneider, D. J. et al. Chapter 8 - Somamer reagents and the somascan platform: Chemically modified aptamers and their applications in therapeutics, diagnostics, and proteomics. In Giangrande, P. H., de Franciscis, V. & Rossi, J. J. (eds.) RNA Therapeutics, 171–260 (Academic Press, 2022).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

