DDX41 coordinates RNA splicing and transcriptional elongation to prevent DNA replication stress in hematopoietic cells
- PMID: 36229594
- PMCID: PMC9613458
- DOI: 10.1038/s41375-022-01708-9
DDX41 coordinates RNA splicing and transcriptional elongation to prevent DNA replication stress in hematopoietic cells
Abstract
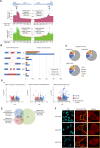
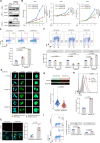
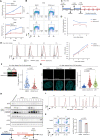
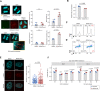
Myeloid malignancies with DDX41 mutations are often associated with bone marrow failure and cytopenia before overt disease manifestation. However, the mechanisms underlying these specific conditions remain elusive. Here, we demonstrate that loss of DDX41 function impairs efficient RNA splicing, resulting in DNA replication stress with excess R-loop formation. Mechanistically, DDX41 binds to the 5' splice site (5'SS) of coding RNA and coordinates RNA splicing and transcriptional elongation; loss of DDX41 prevents splicing-coupled transient pausing of RNA polymerase II at 5'SS, causing aberrant R-loop formation and transcription-replication collisions. Although the degree of DNA replication stress acquired in S phase is small, cells undergo mitosis with under-replicated DNA being remained, resulting in micronuclei formation and significant DNA damage, thus leading to impaired cell proliferation and genomic instability. These processes may be responsible for disease phenotypes associated with DDX41 mutations.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Churpek JE, Smith-Simmer K. DDX41-Associated Familial Myelodysplastic Syndrome and Acute Myeloid Leukemia. In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Gripp KW, et al. editors. University of Washington, Seattle: GeneReviews; 2021. - PubMed
-
- Li P, White T, Xie W, Cui W, Peker D, Zeng G, et al. AML with germline DDX41 variants is a clinicopathologically distinct entity with an indolent clinical course and favorable outcome. Leukemia. 2021;36:664–74. - PubMed
-
- Sébert M, Passet M, Raimbault A, Rahmé R, Raffoux E, Sicre de Fontbrune F, et al. Germline DDX41 mutations define a significant entity within adult MDS/AML patients. Blood. 2019;134:1441–4. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

