Traceless cysteine-linchpin enables precision engineering of lysine in native proteins
- PMID: 36229616
- PMCID: PMC9561114
- DOI: 10.1038/s41467-022-33772-1
Traceless cysteine-linchpin enables precision engineering of lysine in native proteins
Abstract
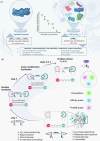
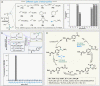
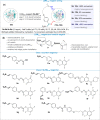
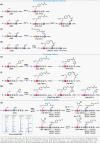
The maintenance of machinery requires its operational understanding and a toolbox for repair. The methods for the precision engineering of native proteins meet a similar requirement in biosystems. Its success hinges on the principles regulating chemical reactions with a protein. Here, we report a technology that delivers high-level control over reactivity, chemoselectivity, site-selectivity, modularity, dual-probe installation, and protein-selectivity. It utilizes cysteine-based chemoselective Linchpin-Directed site-selective Modification of lysine residue in a protein (LDMC-K). The efficiency of the end-user-friendly protocol is evident in quantitative conversions within an hour. A chemically orthogonal C-S bond-formation and bond-dissociation are essential among multiple regulatory attributes. The method offers protein selectivity by targeting a single lysine residue of a single protein in a complex biomolecular mixture. The protocol renders analytically pure single-site probe-engineered protein bioconjugate. Also, it provides access to homogeneous antibody conjugates (AFC and ADC). The LDMC-K-ADC exhibits highly selective anti-proliferative activity towards breast cancer cells.
© 2022. The Author(s).
Conflict of interest statement
V.R. is the founder of Plabeltech Private Limited. A patent application (US 11,149,058 B2; Applicant: IISER Bhopal and DBT; Inventors: Vishal Rai and Srinivasa Rao Adusumalli) has been granted on the LDM. The remaining authors declare no competing interests.
Figures



Similar articles
-
Human Behavior-Inspired Linchpin-Directed Catalysis for Traceless Precision Labeling of Lysine in Native Proteins.Bioconjug Chem. 2022 Dec 21;33(12):2370-2380. doi: 10.1021/acs.bioconjchem.2c00454. Epub 2022 Nov 16. Bioconjug Chem. 2022. PMID: 36383773
-
Linchpin-directed precise labeling of lysine in native proteins, purification, and analysis.Methods Enzymol. 2022;675:383-396. doi: 10.1016/bs.mie.2022.07.016. Epub 2022 Sep 9. Methods Enzymol. 2022. PMID: 36220278
-
Chemoselective and Site-Selective Lysine-Directed Lysine Modification Enables Single-Site Labeling of Native Proteins.Angew Chem Int Ed Engl. 2020 Jun 22;59(26):10332-10336. doi: 10.1002/anie.202000062. Epub 2020 Apr 20. Angew Chem Int Ed Engl. 2020. PMID: 32171045
-
Reactivity and Selectivity Principles in Native Protein Bioconjugation.Chem Rec. 2021 Aug;21(8):1941-1956. doi: 10.1002/tcr.202100108. Epub 2021 Jun 28. Chem Rec. 2021. PMID: 34184826 Review.
-
Site-selective lysine conjugation methods and applications towards antibody-drug conjugates.Chem Commun (Camb). 2021 Oct 14;57(82):10689-10702. doi: 10.1039/d1cc03976h. Chem Commun (Camb). 2021. PMID: 34570125 Free PMC article. Review.
Cited by
-
Disintegrate (DIN) Theory Enabling Precision Engineering of Proteins.ACS Cent Sci. 2023 Jan 23;9(2):137-150. doi: 10.1021/acscentsci.2c01455. eCollection 2023 Feb 22. ACS Cent Sci. 2023. PMID: 36844488 Free PMC article. Review.
-
A simple method for developing lysine targeted covalent protein reagents.Nat Commun. 2023 Dec 1;14(1):7933. doi: 10.1038/s41467-023-42632-5. Nat Commun. 2023. PMID: 38040731 Free PMC article.
-
Multisite Labeling of Proteins Using the Ligand-Directed Reactivity of Triggerable Michael Acceptors.Bioconjug Chem. 2023 Jun 21;34(6):1130-1138. doi: 10.1021/acs.bioconjchem.3c00155. Epub 2023 May 23. Bioconjug Chem. 2023. PMID: 37220065 Free PMC article.
-
Bioinspired one-pot furan-thiol-amine multicomponent reaction for making heterocycles and its applications.Nat Commun. 2023 Jul 10;14(1):4086. doi: 10.1038/s41467-023-39708-7. Nat Commun. 2023. PMID: 37429878 Free PMC article.
-
Efficient Radiolabeling of Proteins and Antibodies via Maleamate-Cysteine Bioconjugation.ACS Med Chem Lett. 2024 Apr 10;15(5):691-695. doi: 10.1021/acsmedchemlett.4c00092. eCollection 2024 May 9. ACS Med Chem Lett. 2024. PMID: 38746876 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

