Co-expression of a PD-L1-specific chimeric switch receptor augments the efficacy and persistence of CAR T cells via the CD70-CD27 axis
- PMID: 36229619
- PMCID: PMC9561169
- DOI: 10.1038/s41467-022-33793-w
Co-expression of a PD-L1-specific chimeric switch receptor augments the efficacy and persistence of CAR T cells via the CD70-CD27 axis
Abstract
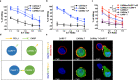
Co-expression of chimeric switch receptors (CSRs) specific for PD-L1 improves the antitumor effects of chimeric antigen receptor (CAR) T cells. However, the effects of trans-recognition between CSRs and PD-L1 expressed by activated CAR T cells remain unclear. Here, we design a CSR specific for PD-L1 (CARP), containing the transmembrane and cytoplasmic signaling domains of CD28 but not the CD3 ζ chain. We show that CARP T cells enhance the antitumor activity of anti-mesothelin CAR (CARMz) T cells in vitro and in vivo. In addition, confocal microscopy indicates that PD-L1 molecules on CARMz T cells accumulate at cell-cell contacts with CARP T cells. Using single-cell RNA-sequencing analysis, we reveal that CARP T cells promote CARMz T cells differentiation into central memory-like T cells, upregulate genes related to Th1 cells, and downregulate Th2-associated cytokines through the CD70-CD27 axis. Moreover, these effects are not restricted to PD-L1, as CAR19 T cells expressing anti-CD19 CSR exhibit similar effects on anti-PSCA CAR T cells with truncated CD19 expression. These findings suggest that target trans-recognition by CSRs on CAR T cells may improve the efficacy and persistence of CAR T cells via the CD70-CD27 axis.
© 2022. The Author(s).
Conflict of interest statement
Z.T. and P.L. are scientific founders of GZI and have equity in GZI. The remaining authors declare no competing interests.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

