An overview of the SAMPL8 host-guest binding challenge
- PMID: 36229622
- PMCID: PMC9596595
- DOI: 10.1007/s10822-022-00462-5
An overview of the SAMPL8 host-guest binding challenge
Abstract
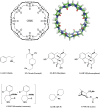
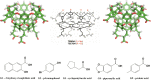
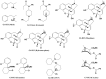
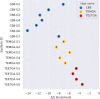
The SAMPL series of challenges aim to focus the community on specific modeling challenges, while testing and hopefully driving progress of computational methods to help guide pharmaceutical drug discovery. In this study, we report on the results of the SAMPL8 host-guest blind challenge for predicting absolute binding affinities. SAMPL8 focused on two host-guest datasets, one involving the cucurbituril CB8 (with a series of common drugs of abuse) and another involving two different Gibb deep-cavity cavitands. The latter dataset involved a previously featured deep cavity cavitand (TEMOA) as well as a new variant (TEETOA), both binding to a series of relatively rigid fragment-like guests. Challenge participants employed a reasonably wide variety of methods, though many of these were based on molecular simulations, and predictive accuracy was mixed. As in some previous SAMPL iterations (SAMPL6 and SAMPL7), we found that one approach to achieve greater accuracy was to apply empirical corrections to the binding free energy predictions, taking advantage of prior data on binding to these hosts. Another approach which performed well was a hybrid MD-based approach with reweighting to a force matched QM potential. In the cavitand challenge, an alchemical method using the AMOEBA-polarizable force field achieved the best success with RMSE less than 1 kcal/mol, while another alchemical approach (ATM/GAFF2-AM1BCC/TIP3P/HREM) had RMSE less than 1.75 kcal/mol. The work discussed here also highlights several important lessons; for example, retrospective studies of reference calculations demonstrate the sensitivity of predicted binding free energies to ethyl group sampling and/or guest starting pose, providing guidance to help improve future studies on these systems.
Keywords: Binding affinity; Blind challenge; Cucurbituril; Free energy; Host–guest binding; Octaacid; SAMPL.
© 2022. The Author(s).
Conflict of interest statement
DLM is a Member of the Scientific Advisory Board of OpenEye Scientific Software, and DLM is an Open Science Fellow with Roivant Therapeutics.
Figures


Similar articles
-
SAMPL7 Host-Guest Challenge Overview: assessing the reliability of polarizable and non-polarizable methods for binding free energy calculations.J Comput Aided Mol Des. 2021 Jan;35(1):1-35. doi: 10.1007/s10822-020-00363-5. Epub 2021 Jan 4. J Comput Aided Mol Des. 2021. PMID: 33392951 Free PMC article.
-
Application of the alchemical transfer and potential of mean force methods to the SAMPL8 host-guest blinded challenge.J Comput Aided Mol Des. 2022 Jan;36(1):63-76. doi: 10.1007/s10822-021-00437-y. Epub 2022 Jan 21. J Comput Aided Mol Des. 2022. PMID: 35059940 Free PMC article.
-
Overview of the SAMPL6 host-guest binding affinity prediction challenge.J Comput Aided Mol Des. 2018 Oct;32(10):937-963. doi: 10.1007/s10822-018-0170-6. Epub 2018 Nov 10. J Comput Aided Mol Des. 2018. PMID: 30415285 Free PMC article.
-
Overview of the SAMPL5 host-guest challenge: Are we doing better?J Comput Aided Mol Des. 2017 Jan;31(1):1-19. doi: 10.1007/s10822-016-9974-4. Epub 2016 Sep 22. J Comput Aided Mol Des. 2017. PMID: 27658802 Free PMC article. Review.
-
The SAMPL4 host-guest blind prediction challenge: an overview.J Comput Aided Mol Des. 2014 Apr;28(4):305-17. doi: 10.1007/s10822-014-9735-1. Epub 2014 Mar 6. J Comput Aided Mol Des. 2014. PMID: 24599514 Free PMC article. Review.
Cited by
-
The temperature-dependence of host-guest binding thermodynamics: experimental and simulation studies.Chem Sci. 2023 Oct 13;14(42):11818-11829. doi: 10.1039/d3sc01975f. eCollection 2023 Nov 1. Chem Sci. 2023. PMID: 37920355 Free PMC article.
-
Potential distribution theory of alchemical transfer.J Chem Phys. 2025 Feb 7;162(5):054106. doi: 10.1063/5.0244918. J Chem Phys. 2025. PMID: 39902686 Free PMC article.
-
The SAMPL9 host-guest blind challenge: an overview of binding free energy predictive accuracy.Phys Chem Chem Phys. 2024 Mar 20;26(12):9207-9225. doi: 10.1039/d3cp05111k. Phys Chem Chem Phys. 2024. PMID: 38444308 Free PMC article.
-
Binding Selectivity Analysis from Alchemical Receptor Hopping and Swapping Free Energy Calculations.J Phys Chem B. 2024 Nov 7;128(44):10841-10852. doi: 10.1021/acs.jpcb.4c05732. Epub 2024 Oct 29. J Phys Chem B. 2024. PMID: 39468848 Free PMC article.
-
Chemical accuracy for ligand-receptor binding Gibbs energies through multi-level SQM/QM calculations.Phys Chem Chem Phys. 2024 Aug 7;26(31):21197-21203. doi: 10.1039/d4cp01529k. Phys Chem Chem Phys. 2024. PMID: 39073067 Free PMC article.
References
-
- Grimme S, Ehrlich S, Goerigk L. Effect of the damping function in dispersion corrected density functional theory. J Comput Chem. 2011;32(7):1456–1465. - PubMed
-
- Goerigk L, Grimme S. Efficient and accurate double-hybrid-meta-GGA density functionals-evaluation with the extended GMTKN30 database for general main group thermochemistry, kinetics, and noncovalent interactions. J Chem Theory Comput. 2011 - PubMed
-
- Abel R (2016) Accelerating drug discovery with free energy calculations. http://www.alchemistry.org/wiki/images/e/eb/Vertex_talk_5_15_2016_clean3.... Accessed 5 Dec 2016
-
- Abel R, Wang L, Harder ED, Berne BJ, Friesner RA. Advancing drug discovery through enhanced free energy calculations. Acc Chem Res. 2017;50(7):1625–1632. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

