Molecular insights into the heat shock proteins of the human parasitic blood fluke Schistosoma mansoni
- PMID: 36229862
- PMCID: PMC9559072
- DOI: 10.1186/s13071-022-05500-7
Molecular insights into the heat shock proteins of the human parasitic blood fluke Schistosoma mansoni
Abstract
Background: Heat shock proteins (HSPs) are evolutionarily conserved proteins, produced by cells in response to hostile environmental conditions, that are vital to organism homeostasis. Here, we undertook the first detailed molecular bioinformatic analysis of these important proteins and mapped their tissue expression in the human parasitic blood fluke, Schistosoma mansoni, one of the causative agents of the neglected tropical disease human schistosomiasis.
Methods: Using bioinformatic tools we classified and phylogenetically analysed HSP family members in schistosomes, and performed transcriptomic, phosphoproteomic, and interactomic analysis of the S. mansoni HSPs. In addition, S. mansoni HSP protein expression was mapped in intact parasites using immunofluorescence.
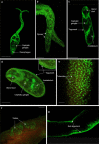
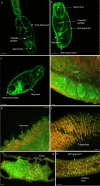
Results: Fifty-five HSPs were identified in S. mansoni across five HSP families; high conservation of HSP sequences were apparent across S. mansoni, Schistosoma haematobium and Schistosoma japonicum, with S. haematobium HSPs showing greater similarity to S. mansoni than those of S. japonicum. For S. mansoni, differential HSP gene expression was evident across the various parasite life stages, supporting varying roles for the HSPs in the different stages, and suggesting that they might confer some degree of protection during life stage transitions. Protein expression patterns of HSPs were visualised in intact S. mansoni cercariae, 3 h and 24 h somules, and adult male and female worms, revealing HSPs in the tegument, cephalic ganglia, tubercles, testes, ovaries as well as other important organs. Analysis of putative HSP protein-protein associations highlighted proteins that are involved in transcription, modification, stability, and ubiquitination; functional enrichment analysis revealed functions for HSP networks in S. mansoni including protein export for HSP 40/70, and FOXO/mTOR signalling for HSP90 networks. Finally, a total of 76 phosphorylation sites were discovered within 17 of the 55 HSPs, with 30 phosphorylation sites being conserved with those of human HSPs, highlighting their likely core functional significance.
Conclusions: This analysis highlights the fascinating biology of S. mansoni HSPs and their likely importance to schistosome function, offering a valuable and novel framework for future physiological investigations into the roles of HSPs in schistosomes, particularly in the context of survival in the host and with the aim of developing novel anti-schistosome therapeutics.
Keywords: Gene expression; Heat shock protein 10; Heat shock protein 40; Heat shock protein 70; Heat shock protein 90; Heat shock proteins; Phosphorylation; Protein expression; Protein–protein association network; Schistosome.
© 2022. The Author(s).
Conflict of interest statement
AJW is a member of the editorial board of Parasites and Vectors. The other authors declare that they have no competing interests.
Figures










References
-
- World Health Organization . Schistosomiasis: control report 2001–2011 and strategic plan 2012–2020. Geneva: WHO Press; 2013.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

